Team:BYU Provo/Team OxyR/Week20
From 2011.igem.org
(Difference between revisions)
Cheddar3210 (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
{{Template:BYU_Provo}} | {{Template:BYU_Provo}} | ||
<html><head><meta http-equiv="X-UA-Compatible" content="IE=EmulateIE7"></head></html> | <html><head><meta http-equiv="X-UA-Compatible" content="IE=EmulateIE7"></head></html> | ||
| - | |||
<div id='pageContents'> | <div id='pageContents'> | ||
| - | + | <html><script> | |
| + | var ele = document.getElementById('liOxyR20'); | ||
| + | ele.style.fontWeight = "bold"; | ||
| + | ele.style.backgroundColor = "#059469"; | ||
| + | jQuery(document).ready(function(){ | ||
| + | $('.accordion').next().show(); | ||
| + | $('#OxyRList').show(); | ||
| + | }); | ||
| + | </script></html> | ||
==30 August 2011== | ==30 August 2011== | ||
Latest revision as of 21:42, 23 September 2011

|
30 August 2011
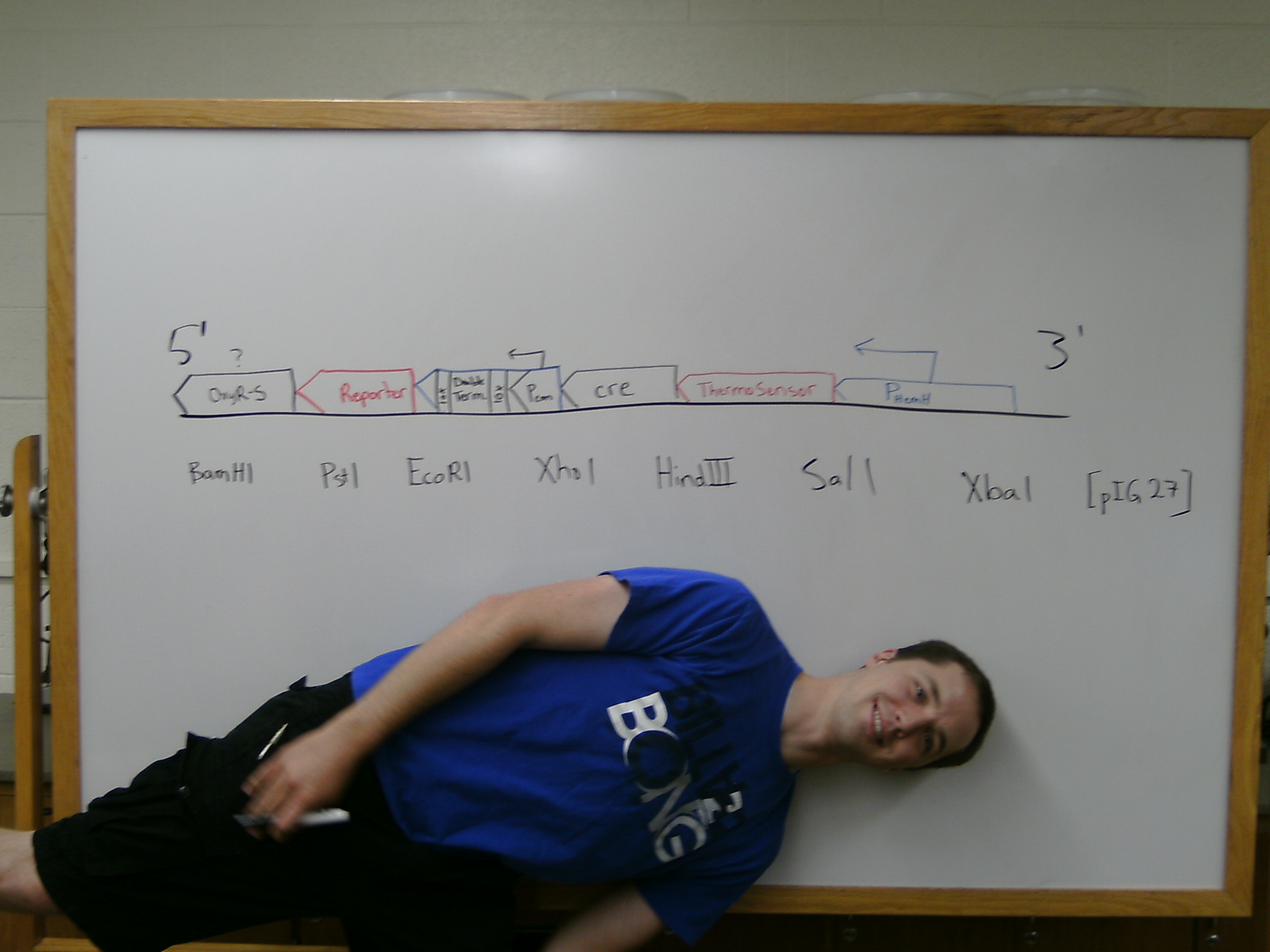
After plasmid minipreps with the new low-copy constructs, Matt electroporated them into the correct wild-type or OxyR- strains.
31 August 2011
Mackay set up a PCR reaction with high-fidelity polymerase, to amplify our katG and hemH promoters with new restriction sites, in order to clone them into the thermosensor team's construct with minimal work. Matt came in later that night to set up overnights for the next day's plate reader experiments.
1 September 2011
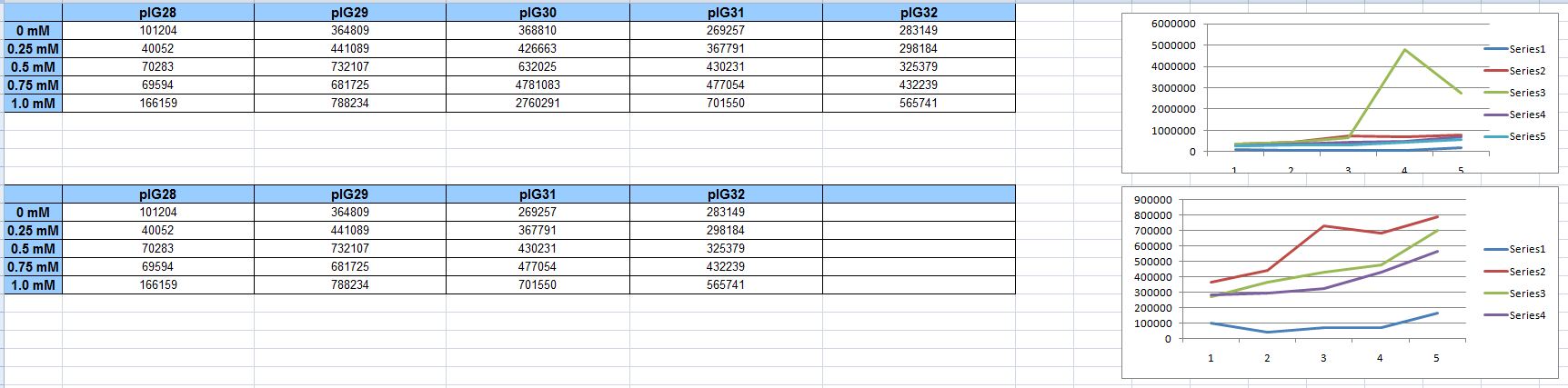
Matt and Mackay were in the lab bright and early to run the plate reader experiment with the new low-copy constructs, and with their minds made up to make no mistakes whatsoever. Indeed, it was basically a flawless execution. All constructs were run in triplicate. The data looked great:
 "
"