Team:BYU Provo/Team OxyR/The Beginning
From 2011.igem.org

|
5 January 2011
Colon Cancer Detection Questions
Can isopeptags be used to detect a cancer biomarker and then create a response inside the cell? If not, what other transmembrane proteins can?
Heat Production
[http://openwetware.org/images/f/fd/Cold_Switch_Mechanism.jpg Diagram of proposed mechanism]
What's a good "cold switch" - a riboswitch that would stop transcription at a specific temperature?
Does the UCP1 create enough heat, fast enough, to make a difference?
Does activation of UCP1 harmfully inhibit ATP production? Or can we overproduce ATP to compensate?
CO Detection
Can E. coli be altered to survive in CO long enough to detect it?
7 January 2011
Heat Production
We discovered that the yeast heating project was originally done by an iGEM team from Valencia. If we continue with it, our original contribution would be linking it to a riboswitch, and optimizing the heating mechanism. Addison also suggested optimizing it in E. coli because they grow in denser colonies (it has already been shown not to be lethal in E. coli, but we aren't sure if it produces heat).
Furthermore, there is a better chance of E. coli heating its environment more efficiently as the UCP-1 protein would be expressed on its membrane as opposed to the mitochondrial membrane.
Pamphlet printing is cheap, and Rob volunteered to work on the graphic design.
Colon Cancer Detection
We've found that cancer cells produce more reactive oxygen species than normal cells. E. Coli already has detection systems for ROS so we could couple those detection systems with some sort of report to detect the ROS. Such as GFP for fluorescence, or banana aroma for a little added flavor.
The carbon monoxide project ...
10 January 2011
We discovered that the CO detection project was already done by an iGEM team last year.
Brainstorming:
- Sound-sensitive bacteria using TRP channels or MSCL channel
- Engineer Lactobascillus to add vitamins to yogurt
- ROS sensor in E. coli to detect cancer (and/or heat sensitivity)
- Thermogenesis in E. coli
12 January 2011
Continued Brainstorming
- Caffeine synthesis (Starbacts :)
- Creating a psychrophile (cold-loving) E. coli (E. skimocoli :) using phychrophilic proteins, chaperonins and UCP1
- Pathogenic plasmid-sharing bacteria (shares lactase-encoding plasmid with other flora in gut)
- Fabric (long bacteria that interlace ... create polymer within cell then allow to die ... when cell dies, long polymers are already interlaced)
- Paper (decompose plant matter and create pulp that can be made into paper)
- Sound detection (TRP proteins, MSCL + MSCS proteins in bacteria)
- Organic nanowires
- Electron sharing between species
- Cell migration
We settled on Cancer detection, when all was said and done...
Colon Cancer Detection Mechanism Proposals for Continued Research
The idea is to detect various inputs and create unique reporters or levels of reporters according to inputs. To detect colon cancer we would focus on two or more of the following:
- Unique metabolic products (Fermentation - lactic acids)
- Reactive Oxygen Species (ROS)
- Heat Detection (Riboswitch)
- Collagen cutting enzymes (detect the byproducts)
Unique parts will include: ROS sensor (fine-tune those existing in nature), Riboswitch sensitive to specific temperature, and multi-input switch (creating our own).
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K228258 Sample AND switch]
Pamphlet Design
Trifold Pamphlet:
- Who we are
- What is iGEM
- Our Goals within iGEM
- What we need
- How you can help
14 January 2011
Wrote pamphlet text. See Papers section.
19 January 2011
Proofread pamphlet text and plan to take pictures on Friday. Friday afternoon, Addison and his friend will work on the layout.
Started research in earnest for colon cancer biomarkers that will be useful in our project.
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WN2-4NRMDCT-3&_user=456938&_coverDate=12%2F31%2F2007&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_searchStrId=1613028817&_rerunOrigin=google&_acct=C000021830&_version=1&_urlVersion=0&_userid=456938&md5=6434a6e2cbf35e44e5540c2b1799759c&searchtype=a ROS are decreased in cancerous cells]
[http://www.ncbi.nlm.nih.gov/pubmed/1331990 ROS are increased in CRC]
ROS-sensing transcriptional regulators already in E. Coli are OxyR, SoxR, and FNR. These could potentially be used to detect different concentrations of ROS.
Biomarkers Unique to Colon Cancer
[http://en.wikipedia.org/wiki/Carcinoembryonic_antigen Carcinoembryonic antigen]
[http://cebp.aacrjournals.org/content/12/10/1095.full.pdf+html Detection of Glycoprotein 87 Using Monoclonal Antibody Adnab-9]
[http://cebp.aacrjournals.org/content/17/3/543.full.pdf+html Consistently Reported Candidate Biomarkers - Check last pages for lists of genes/proteins]
[http://www.aolnews.com/2010/03/22/scientists-find-first-ever-colon-cancer-biomarkers/ TFF3 and GDF15]
Goals for Next Few Weeks
Subjects to Study: Circuit design Directed evolution Mathematical modeling Colon cancer biology basics Other tools that will be useful.
Professors to give lectures: Dr. Clement Dr. Grose Dr. O'Neil Dr. Griffitts
21 January 2011
Assignments: Matt and Rob - ROS Transcription Factors
Addison and Chet - Riboswitches
Mark and Cameron (and Josh?) - Colon Cancer Specifics (Where are biomarkers released? Where and how does it develop, etc.)
Mackay and Julie - AND/OR Gate mechanisms [http://openwetware.org/images/9/95/AND_GateBYU.jpg AND Gate Mechanism]
GOAL: Detailed strategy and schedule within two weeks
24 January 2011
- Can internal biomarkers be used by our E. coli if the cells die and are found in the stool?
- What biomarkers are common to most colon cancers?
- What are the stages of cancer development and how much do signals/biomarkers change through the stages?
- ROS/ROM's ... do they reach the environment? What causes them? Are they a good signal?
- Anatomy and pathophysiology of the cancer? What is a polyp? Where does it dump things out? Is a polyp made only of cancer, or is it made of mixed cells?
And Or Gate Switches
Various Gates-AND,OR,NAND,XOR,NOT Various promoters How to get best combined response from two inputs
Colon Cancer
Colon cancer begins as normal colonic epithelium which can develop into an adenomatous polyp which can further develop in to an invasive cancer polyp-abnormal growth of tissue projecting from a mucous membrane Polyps are an early warning for colon cancer. There are two types of colon cancer: 1.adenoma-pre malignant polyp 2.hyperplastic polyps-rarely forms a cancerous tumor
26 January 2011
Mathematical Modeling Example (UBC Example)
28 January 2011
Circuit Design Brainstorming
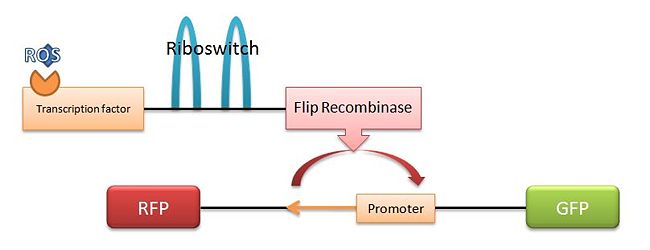
We spent the day coming up with various mechanisms to express a desired gene in the presence of ROS and heat. Once expressed properly, we would like our marker gene of choice to be expressed constitutively. Following are four proposed mechanisms. The mechanism we have decided to try is the fourth mechanism, which we have named the "SuperFlp Mechanism." Below is a diagram.
- Basic FLp Recombinase Mechanism: A DNA section that is transcribed normally in the presence of ROS, but there is nothing important downstream. When heated, the promoter inverts (5' to 3' as well as switching strands), leading to transcription in the opposite direction, which contains our desired marker.
- IRES Mechanism: The promoter on the strand is ROS activated, but the mRNA lacks a Shine Dalgarno sequence. The SD sequence will be inserted by a heat-activated internal ribosomal entry site (IRES) in the proper position to result in translation of the marker protein.
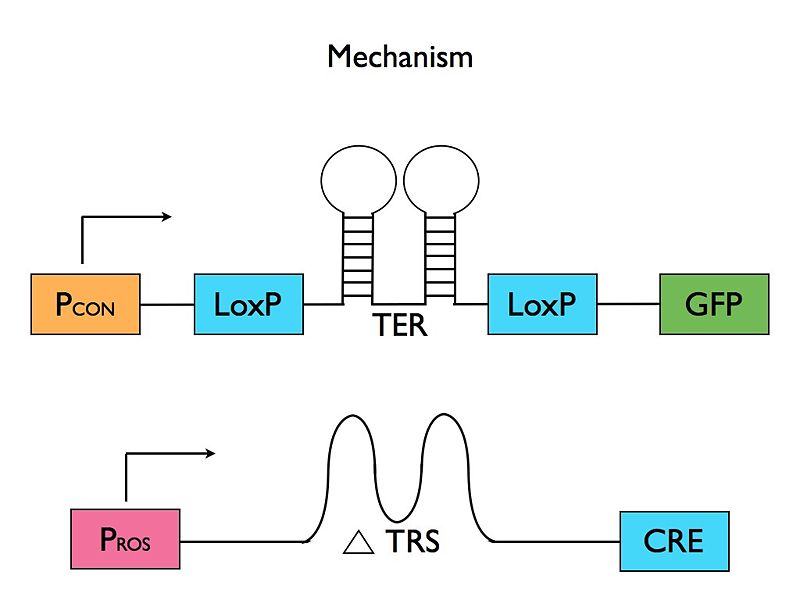
- Riboswitch Mechanism: The presence of ROS leads to translation of protein A. Protein A is required for translation of the desired mRNA coding for the marker. The mRNA starts with a heat-activated riboswitch flanked by LoxP sites and followed by the Cre protein. So, when activated by heat, the heat-sensitive riboswitch will be eliminated, allowing constituent translation. The Cre sequence is followed by a duplicate sequence for protein A.
- SuperFlp Mechanism: (Method of choice) An ROS transcription factor is followed by a riboswitch and a Flp recombinase. In the presence of both ROS and heat, the recombinase is expressed, flipping non-local promoter to the desired position. The big question is whether the Flp recombinase's work will be undone by time or the same Flp recombinase. If the recombination is permanent, the E. Coli will continue to express the desired marker even when no longer in the presence of ROS or heat.
January 31 2011
Normal ROS levels in E. coli
Hydrogen peroxide:
"The activity of alkylhydroperoxide reductase (Ahp) is so high that, although H2O2 is formed endogenously inside aerobic E. coli at a rate of about 15 μM/s, the steady-state concentration does not exceed 20 nM."
"When these higher doses of H2O2 saturate Ahp, the H2O2 concentration rises beyond the 0.1 μM threshold for OxyR activation (48, 58). Catalase is strongly induced, and it becomes the primary scavenging enzyme. The katG-encoded catalase of E. coli exhibits a high Km, so it is not saturated by even millimolar doses of H2O2 (60). Importantly, because catalase dismutates H2O2, its turnover rate is not restricted by the availability of reducing equivalents."
Superoxide:
"Exponentially growing E. coli contains about 20 μM dimeric cytoplasmic SOD. Given that the rate of O2− formation is approximately 5 μM/s in these cells, the outcome is that the steady-state level of O2− is restricted to approximately 0.1 nM (53)."
Source: [http://www.annualreviews.org/doi/full/10.1146/annurev.biochem.77.061606.161055 Cellular Defenses against Superoxide and Hydrogen Peroxide]
Diagram of the final mechanism
Links to Gene Sequences
[http://www.ncbi.nlm.nih.gov/nuccore/NC_003210?report=genbank&from=2619415&to=2620491&strand=true prfa Gene in listeria (should contain thermoswitch)]
[http://sfx.lib.byu.edu/sfxlcl3?url_ver=Z39.88-2004;url_ctx_fmt=info:ofi/fmt:kev:mtx:ctx;rft_val_fmt=info:ofi/fmt:kev:mtx:journal;rft.atitle=An%20RNA%20thermosensor%20controls%20expression%20of%20virulence%20genes%20in%20Listeria%20monocytogenes;rft.auinit=J;rft.aulast=Johansson;rft.date=2002;rft.epage=561;rft.genre=article;rft.issn=0092-8674;rft.issue=5;rft.spage=551;rft.stitle=CELL;rft.title=CELL;rft.volume=110;rfr_id=info:sid/www.isinet.com:WoK:WOS;rft.au=Mandin,%20P;rft.au=Renzoni,%20A;rft.au=Chiaruttini,%20C;rft.au=Springer,%20M;rft.au=Cossart,%20P An RNA Thermosensor Controls Expression of Virulence Genes in Listeria monocytogenes]
[http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG10681 OxyR Information]
February 2, 2011
Thermoswitch
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WSN-4C5HD56-4&_user=456938&_coverDate=09%2F06%2F2002&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_acct=C000021830&_version=1&_urlVersion=0&_userid=456938&md5=a085040d8d9c38e71c715f579a400156&searchtype=a The Johansson Paper]
OxyR
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC177537/pdf/1776740.pdf?tool=pmcentrez Mapping of the OxyR Protein Contact Site in the C-Terminal Region of RNA Polymerase a Subunit] Says that transcription goes up only 2.73 fold in presence of H2O2. Need to double check this.
[http://jb.asm.org/cgi/content/full/181/9/2759 In Vivo Transcription of the Escherichia coli oxyR Regulon as a Function of Growth Phase and in Response to Oxidative Stress] Says that genes controlled by OxyR only increase about 11-fold when in the presence of H2O2.
[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T4P-3Y0HP9V-1&_user=456938&_coverDate=01/01/2000&_rdoc=1&_fmt=high&_orig=search&_origin=search&_sort=d&_docanchor=&view=c&_searchStrId=1628582425&_rerunOrigin=google&_acct=C000021830&_version=1&_urlVersion=0&_userid=456938&md5=327115394eca4ffc8274fbdcc3b31ae5&searchtype=a Redox sensing by prokaryotic transcription factors] Says that SoxR increases transcription by 100-fold.
February 4, 2011
Thermoswitch Team
[http://rna.urmc.rochester.edu/RNAstructureDownload.html RNA Structure Program]
Multiple cloning site addresses
Brief idea of how we want to arrange everything in the multiple cloning site.
OxyR and SoxR work
Designed primers to PCR the E. Coli genes.
Playing with thermoswitches on our computer program
Of the sequences listed below, we decided to use Mutation11, GriffShort, GriffMed, GriffLong, and the original listeria sequence. Sequences were picked based on their similarity to the original listeria sequence either in folding conformation or in energy prediction.
Original sequence taken from the listeria paper: UGUAAAAAACAUCAUUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUUACUGCCUAAUG UUUUUAGGGUAUUUUAAAAAAGGGCGAUAAAAAACGAUUGGGGGAUGAGACAUGAACGCUCAAGCA Energy: -23.5
Mutation1 UGUAAAAAACAUCAUUUAGCGUGACUUUCCAAUUUCAGCUAACAAUUGUUGUUACUGCCUAAUGU UUUUAGGGUAUUUUAAAAAAGGGCGAUAAAAAACGAUUGGGGGAUGAGACAUGAACGCUCAAGCA Energy:-25.4
MutationSD UGUAAAAAACAUCAUUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUUACUGCCUAAUGU UUUUAGGGUAUUUUAAAAAAGGGCGAUAAAAAACGAUUGGAGGAGGUUGAGACAUGAACGCUCAAGCA Energy:-31.8
MutationSD2: UGUAAAAAACAUCAUUUAGCGUGACUUUCUUUCAACAGGGAACAAUUGUUGUUACUGCCUAAU GUUUUUAGGGUAUUUUAAAAAAGGGCGAUAAAAAACGAUUGGAGGAGGUUGAGACAUGAACGCUCAAGCA Energy:-27.9
Mutation GriffLong: Cut 14nt from 5' and ant from 3'; Changed SD to GGAGG from GGGGG. UUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUUACUGCCUAAUGUUUUUAGGGUA UUUUAAAAAAGGGCGAUAAAAAACGAUUGGAGGAUGAGACAUGAACGCUCAA Energy:-24.1
Mutation GriffLong+: Added the correct 2 bases back onto the GriffLong. CAUUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUUACUGCCUAAUGUUUUUAGGG UAUUUUAAAAAAGGGCGAUAAAAAACGAUUGGAGGAUGAGACAUGAACGCUCAAGC Energy:-24.1
Mutation GriffMed: Cut a sequence to shorten the loops. UUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUUACUGCCUAAUGUAAGGGCGAUA AAAAACGAUUGGAGGAUGAGACAUGAACGCUCAA Energy:-22.9
Mutation GriffShort: Cut another sequence to shorten. UUUAGCGUGACUUUCUUUCAACAGCUAACAAUUGUUGUAAAAACGAUUGGAGGAUGAGAC AUGAACGCUCAA Energy:-16.3
Mutation GriffShort2: Changed some bases to increase pairing. UUUAGCGUUCCUUUCUUUCAACAGCUAACAAUUGUUGUAAAAACGAUUGGAGGAUGAGA CAUGAACGCUCAA Energy:-19.8
Mutation9: Continued base changes to increase pairing. UUUAGCGUUCCUUUCUUUCAACAGCUAACAAUUGUUGUAAAAACGAUUGGAGGAUGAG ACAUGAACGCUCAA Energy:-17.7
Mutation10: UUUAGCGCUCCUUUCUUUCAACAGCUAACAAUUGUUGUAAAAACGAUUGGAGGAUGA GACAUGAGCGCUCAA Energy:-22.2
Mutation11: UUUGGCGCUCCUUUCUUUCAACAGCUAACAAUUGUUGUAAAAACGAUUGGAGGAUGAG ACAUGAGCGCCCAA Energy:-24.2
9 February 2011
Multiple Cloning Site Addresses
Updated idea of our multiple cloning sites.
Team ΔTRS
- Made small primers (F & R) for GFP with restriction sites for Pbad
- Made long primers (F & R) for five variations of our ΔTRS.
11 February 2011
Thermoswitch Team
We've designed our forward and reverse primers to clone out the ara-C gene and pBAD promoter for our thermoswitch plasmid.
ara-C pBAD Forward Primer:
gccAGATCTtaccgggtagaatcaaaccga
ara-C pBAD Reverse Primer: cggCTGCAGaaaaacgggTatggagaaaca
Oxy Team
Redesigned the multiple cloning site (MCS) map in light of some new information regarding the OxyR binding site. Click image below to see details.
OxySoxy Team To Do:
- Check PCR
- if good, restriction enzyme digest
- if not good, PCR again
- Plasmid:
- extract from E. coli
- PCR amplification
ΔTS Team To Do:
- PCR amplification of primers
- Plasmid extraction
- Insertion
16 February 2011
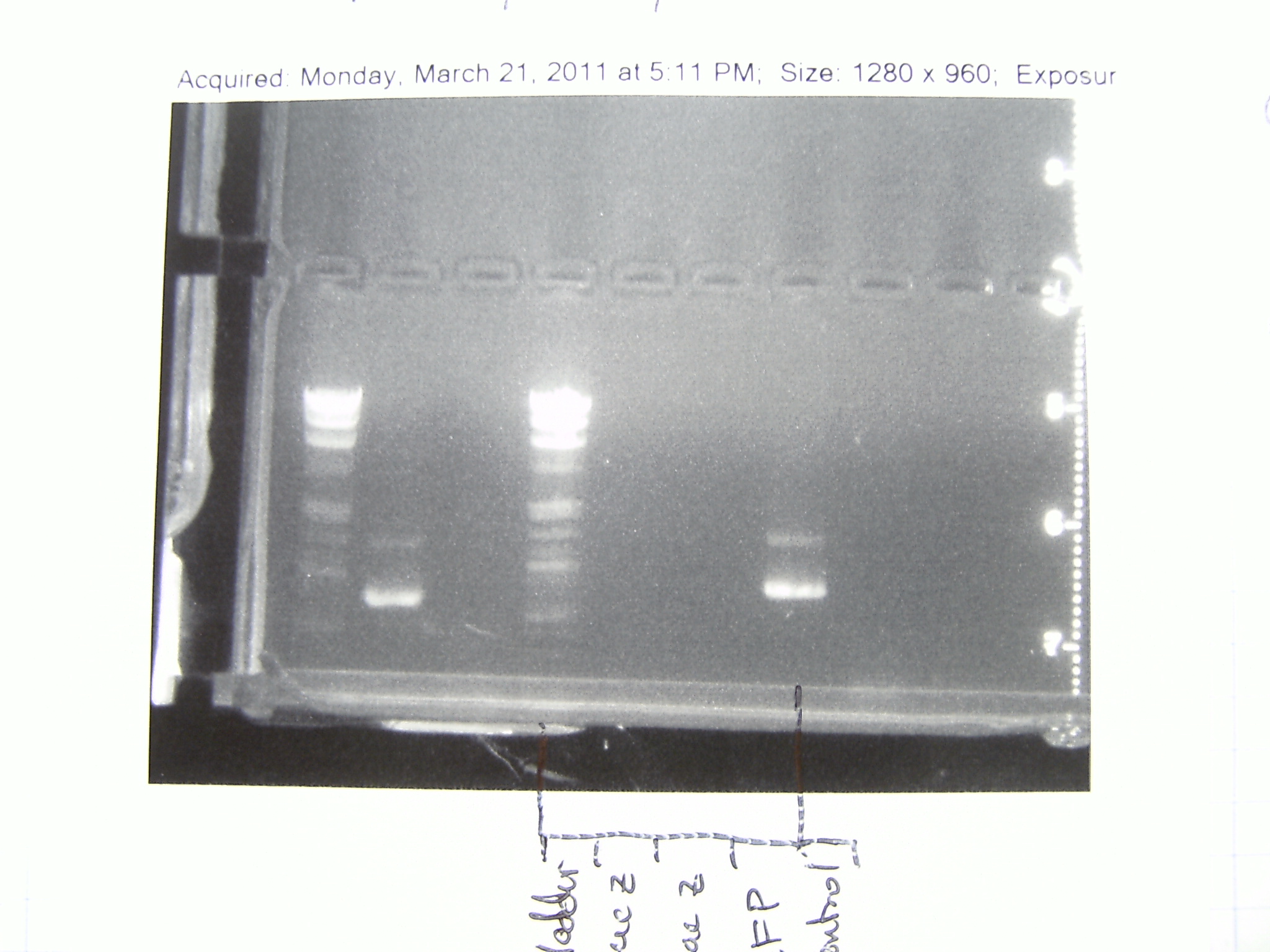
We checked the PCR we ran on Monday and there were only faint bands for OxyR-Weak and OxyR-Medium. We set up the PCR again and hope it works better this time.
18 February 2011
Gel picture of the PCR we ran on the 16th. Looks great!
23 February 2011
Ran a miniprep using standard protocol.
10 March 2011
Ran PCR to amplify GFP, to clone into pPlat. The primers added an XbaI and EcoRI site to the ends of the GFP fragment.
21 March 2011
On Wednesday last week we did the restriction enzyme digest using XbaI on the pPlat plasmid and the GFP with both forward and reverse primers containing XbaI (a non-directional cloning). There was some pipetting error that diluted the solution. We ran it on a low-melt gel and cut out the bands.
Today we ligated the plasmid(pPlat) and the GFP, and transformed into DH5alpha. Addison showed us how to plate the bacteria and we set it to incubate overnight at 37 degrees overnight. Tomorrow we'll put it in the fridge and continue work on Wednesday.
23 March 2011
Mackay purified the PCR from Monday. We proceeded to digest the GFP insert and the plasmid backbone (pPlat) and prepare them for directional cloning. We also performed colony PCR on the non-directional cloning from March 21.
25 March 2011
We ran the colony PCR's from 23 March on a gel. More than half of the non-directional clonings worked!
28 March 2011
Moved colonies from directional GFP cloning to patches. Also set up another colony PCR to check the inserts in that nondirectional cloning.
30 March 2011
Checked colony PCR from 28 March on a gel. Only once colony had the GFP, but that's enough.
6 April 2011
We purified the directional GFP insert plasmid, ran PCR on the multiple cloning site (MCS) and submitting it for sequencing. The GFP insert was perfect, with no mutations, insertions or deletions.
11 April 2011
Mackay did the PCR with the primers on this gel (these serve as templates for each other ... they prime each other to form the OxyS binding site with sticky ends for the ligation step). They worked, according to Dr. Grose. Because the PCR product is so small, Dr. Grose recommended we suspend the DNA in ethanol and spin it down, rather than use a purification kit.
13 April 2011
Restriction digests from last week were left in the 37 degree heating plate for days. Dr. Grose said they may still have worked fine. We ran it on a low-melt gel but only the plasmid showed up ... not our insert. We reviewed the primers we'd used and decided that they probably require template ... we'd designed the primers badly. We redesigned the primers to anneal to the E. coli SoxS, OxyS, KatG and HemH promoters.
14 April 2011
OxyR team came in on reading day and purified the plasmid with the GFP insert since we forgot to add ampicillin to the overnight culture last time. We boiled some E. coli to use as template for colony PCR to check the binding domain.
 "
"