Team:UNICAMP-EMSE Brazil/Notebook/17 September 2011
From 2011.igem.org

| Home | Project | Methods | Results | Data | Team | Notebook | Human Practices | Safety | Profile | Sponsors | Wix |
Contents |
Notebook
Click on a date to see what we have done!
|
|
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
17 September 2011
Sequences to ship
Quantification and checking digestion of sequences to ship
Objectives:
- Quantification of linearized pSB1C3 vector purified from the gel (volume of ladder applied 6 uL; sample: 5 uL DNA +1 uL buffer),
- Checking digestion ( EcoRI and PstI) of samples mini-preped 09/15 and 09/16.
- Checking digestion of SoxS+HlyB+HlyD +TolC+T C1 1/ SoxS+HlyB+HlyD +TolC+T C1 2/ SoxS+HlyB+HlyD +TolC+T C1 3 (EcoRI and PstI) of samples mini-preped 09/08.
Samples:
- Gel 1:
- pSB1/ pSB 2/ pSB 3/ pSB 4/ pSB 5/ RBS+IL-12 C4/ RBS+IL-12 C5/ RBS+HlyD C4/ RBS+HlyD C5/ RBS+Tol C C10/ RBS+Tol C C11/ RBS+QseB+RBS+QseC C10/ RBS+QseB+RBS+QseC C11/ RBS+QseB+RBS+QseC C12/
- Gel 2:
- RBS+SoxS C10/ RBS+SoxS C11/ RBS+SoxS C12/ RBS+SoxS C13/ RBS+HlyB C10/ RBS+HlyB C11/ RBS+HlyB C12/ SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1/ SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C1 2/ SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C3/
- Ladder:
- 100-10.000 bp (Fermentas)
- Gel Agarose concentration:
- 1,0%
- Results:
| Construction | Total size (gene+vector) | Linear vector size | Gene size | Result |
|---|---|---|---|---|
| pSB 1 | 2070 | 2070 | ---- | Small concentration, ~ 3 ng/uL |
| pSB 2 | 2070 | 2070 | ---- | Small concentration ~ 3 ng/uL |
| pSB 3 | 2070 | 2070 | ---- | Small concentration ~ 3 ng/uL |
| pSB 4 | 2070 | 2070 | ---- | Small concentration ~ 3 ng/uL |
| pSB 5 | 2070 | 2070 | ---- | Small concentration ~ 3 ng/uL |
| RBS+IL-12 C4 | 3728 | 2070 | 1658 | Linearized vector OK. No fragment band. |
| RBS+IL-12 C5 | 3728 | 2070 | 1658 | Linearized vector OK. No fragment band. |
| RBS+HlyD C4 | 3566 | 2070 | 1496 | Linearized vector OK. No fragment band. |
| RBS+HlyD C5 | 3566 | 2070 | 1496 | Linearized vector OK. No fragment band. |
| RBS+Tol C C10 | 3650 | 2070 | 1580 | Linearized vector OK. No fragment band. |
| RBS+Tol C C11 | 3650 | 2070 | 1580 | Linearized vector OK. No fragment band. |
| RBS+QseB+RBS+QseC C10 | 4156 | 2070 | 2086 | Linearized vector OK. No fragment band. |
| RBS+QseB+RBS+QseC C11 | 4156 | 2070 | 2086 | Digestion OK. Strange size for fragment band (maybe it is the closed vector band). |
| RBS+QseB+RBS+QseC C12 | 4156 | 2070 | 2086 | Linearized vector OK. No fragment band. |
| RBS+SoxS C10 | 2173 | 2070 | 103 | Linearized vector OK. No fragment band. |
| RBS+SoxS C11 | 2173 | 2070 | 103 | Linearized vector OK. No fragment band. |
| RBS+SoxS C12 | 2173 | 2070 | 103 | Linearized vector OK. No fragment band. |
| RBS+SoxS C13 | 2173 | 2070 | 103 | Linearized vector OK. No fragment band. |
| RBS+HlyB C10 | 4253 | 2070 | 2183 | Linearized vector OK. Closed vector band. |
| RBS+HlyB C11 | 4253 | 2070 | 2183 | Linearized vector OK. No fragment band. |
| RBS+HlyB C12 | 4253 | 2070 | 2183 | Linearized vector OK. Closed vector band. |
| SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C1 1 | 8662 | 3205 | 5457 | Digestion did not work |
| SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C1 2 | 8662 | 3205 | 5457 | Digestion did not work |
| SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C3 | 8662 | 3205 | 5457 | Digestion OK. Fragment band visible. Closed vector band. |
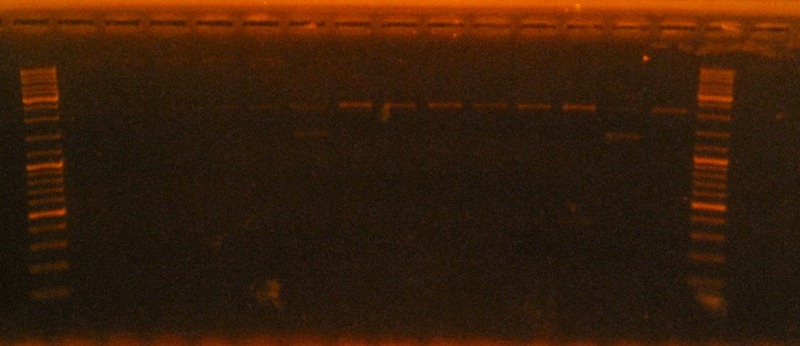
Electrophoresis Gel 3, 17/09/2011.
Gel 3: (d)RBS+HlyD C4/ (nd)RBS+HlyD C4/ (d)RBS+Tol C C11/ (nd)RBS+Tol C C11/ (d)RBS+QseB+RBS+QseC C10/ (nd)RBS+QseB+RBS+QseC C10/(d)RBS+SoxS C13/ (nd)RBS+SoxS C13/ (d)RBS+HlyB C10/ (nd)RBS+HlyB C10/ (d)RBS+HlyB C11/(nd) RBS+HlyB C11/ (d)SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1/(nd) SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1
Gel 3: (d)RBS+HlyD C4/ (nd)RBS+HlyD C4/ (d)RBS+Tol C C11/ (nd)RBS+Tol C C11/ (d)RBS+QseB+RBS+QseC C10/ (nd)RBS+QseB+RBS+QseC C10/(d)RBS+SoxS C13/ (nd)RBS+SoxS C13/ (d)RBS+HlyB C10/ (nd)RBS+HlyB C10/ (d)RBS+HlyB C11/(nd) RBS+HlyB C11/ (d)SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1/(nd) SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1
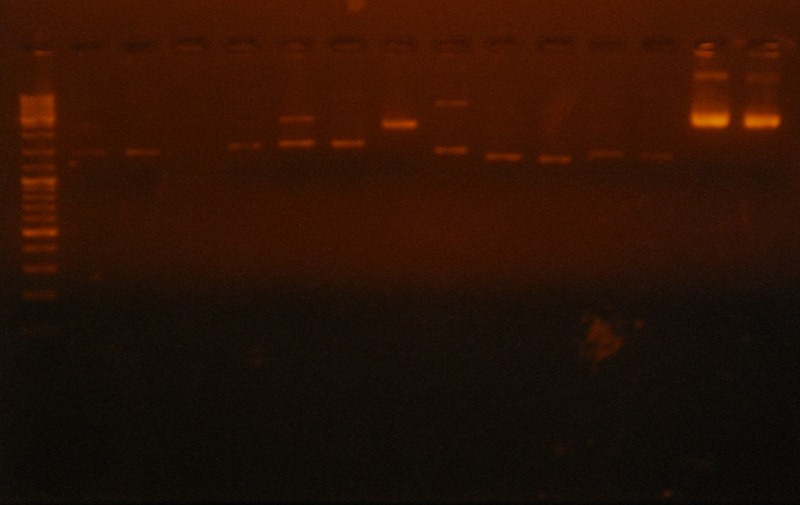
Revising constructs
Objective:
- Checking digestion with Eco RI of some constructs that had strange result in former electrophoresis
Samples:
- Gel 3:
- (d)RBS+HlyD C4/ (nd)RBS+HlyD C4/ (d)RBS+Tol C C11/ (nd)RBS+Tol C C11/ (d)RBS+QseB+RBS+QseC C10/ (nd)RBS+QseB+RBS+QseC C10/(d)RBS+SoxS C13/ (nd)RBS+SoxS C13/ (d)RBS+HlyB C10/ (nd)RBS+HlyB C10/ (d)RBS+HlyB C11/(nd) RBS+HlyB C11/ (d)SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1/(nd) SoxS+ RBS+HlyB +RBS+HlyD+ RBS+TolC+T C1 1
- Results:
| Construction | Total size (gene+vector) | Linear vector size | Gene size | Result |
|---|---|---|---|---|
| (d)RBS+HlyD C4 | 3566 | 2070 | 1496 | Digestion did not work. (Closed vector with same size linearized vector). |
| (nd)RBS+HlyD C4 | 3566 | 2070 | 1496 | Closed vector band ok. |
| (d)RBS+Tol C C11 | 3650 | 2070 | 1580 | ---- |
| (nd)RBS+Tol C C11 | 3650 | 2070 | 1580 | ---- |
| (d)RBS+QseB+RBS+QseC C10 | 4156 | 2070 | 2086 | Digestion did not work. (Closed vector with same size linearized vector). |
| (nd)RBS+QseB+RBS+QseC C10 | 4156 | 2070 | 2086 | Closed vector band ok. |
| (d)RBS+SoxS C13 | 2173 | 2070 | 103 | It seems that ligation is ok, fragment size band matches. |
| (nd)RBS+SoxS C13 | 2173 | 2070 | 103 | Closed vector band ok. |
| (d)RBS+HlyB C10 | 4253 | 2070 | 2183 | Digestion did not work. (Closed vector with same size linearized vector). |
| (nd)RBS+HlyB C10 | 4253 | 2070 | 2183 | Closed vector band ok. |
| (d)RBS+HlyB C11 | 4253 | 2070 | 2183 | Digestion did not work. (Closed vector with same size linearized vector). |
| (nd)RBS+HlyB C11 | 4253 | 2070 | 2183 | Closed vector band ok. |
| (d)SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C1 1 | 8662 | 3205 | 5457 | Digestion did not work |
| (nd)SoxS+ RBS+HlyB+ RBS+HlyD + RBS+TolC+T C1 1 | 8662 | 3205 | 5457 | Closed vector band ok. |
Building the devices
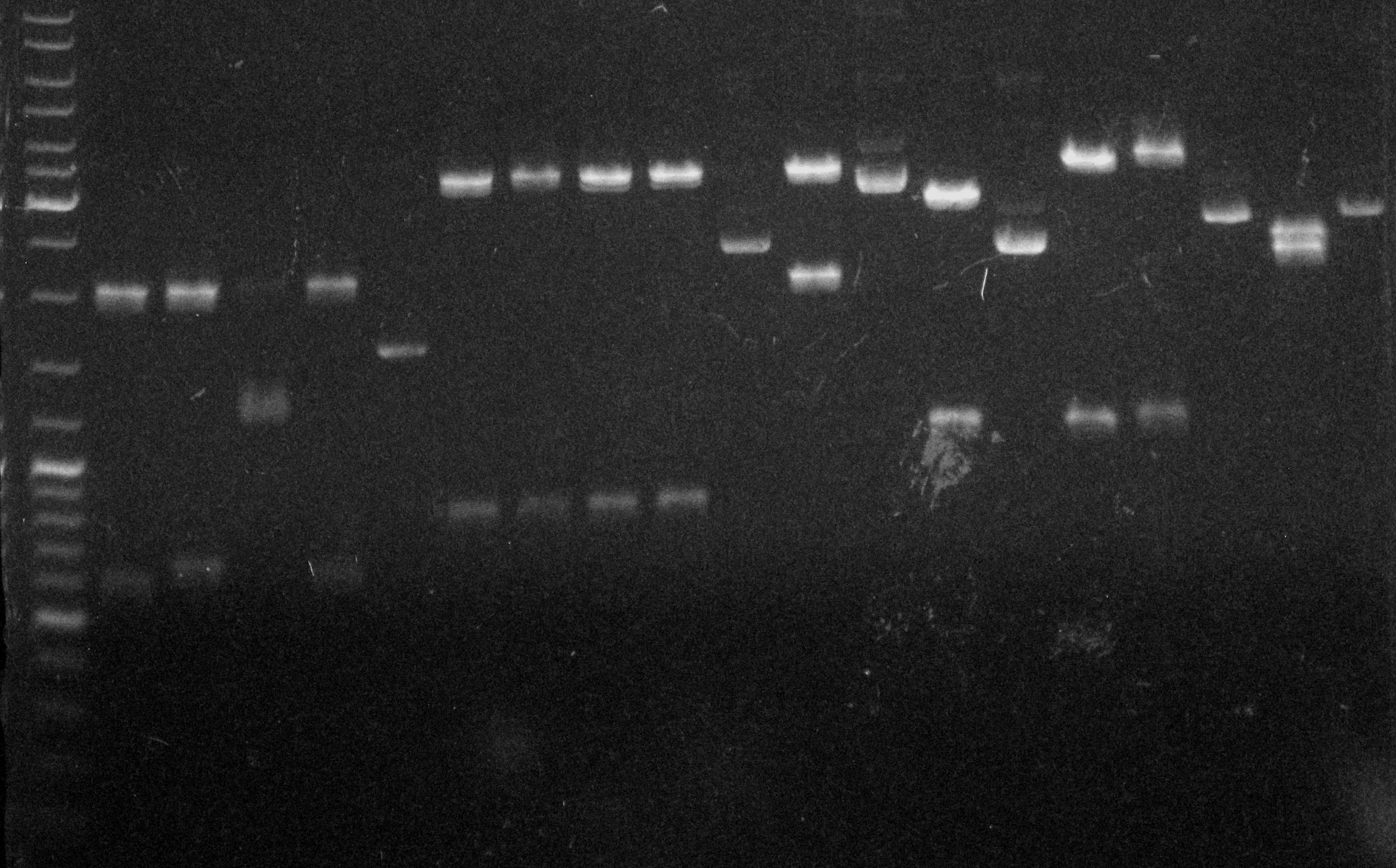
Electrophoresis Gel 4, 17/09/2011.
Gel 4: Ladder / pConst+RBS+SoxR+T C1 (D)/ pConst+RBS+SoxR+T C2 (D)/ pConst+RBS+SoxR+T C3 (D)/ pConst+RBS+SoxR+T C4 (D)/ pConst+RBS+SoxR+T (ND) / SoxS+RBS+IL-10+HlyA+T C1 (D)/ SoxS+RBS+IL-10+HlyA+T C2 (D)/ SoxS+RBS+IL-10+HlyA+T C3 (D)/ SoxS+RBS+IL-10+HlyA+T C4 (D)/ SoxS+RBS+IL-10+HlyA+T (ND)/ flhDC+RBS+IL-12+HlyA+T (D) / flhDC+RBS+IL-12+HlyA+T (ND)/ flhDC+RBS+GFP+HlyA+T (D)/ flhDC+RBS+GFP+HlyA+T (ND)/ SoxS+RBS+GFP+HlyA+T (D)/ SoxS+RBS+GFP+HlyA+T Test (D)/ SoxS+RBS+GFP+HlyA+T (ND) / pConst+RBS+QseB+RBS+QseC+T (D)/ pConst+RBS+QseB+RBS+QseC+T (ND)
Gel 4: Ladder / pConst+RBS+SoxR+T C1 (D)/ pConst+RBS+SoxR+T C2 (D)/ pConst+RBS+SoxR+T C3 (D)/ pConst+RBS+SoxR+T C4 (D)/ pConst+RBS+SoxR+T (ND) / SoxS+RBS+IL-10+HlyA+T C1 (D)/ SoxS+RBS+IL-10+HlyA+T C2 (D)/ SoxS+RBS+IL-10+HlyA+T C3 (D)/ SoxS+RBS+IL-10+HlyA+T C4 (D)/ SoxS+RBS+IL-10+HlyA+T (ND)/ flhDC+RBS+IL-12+HlyA+T (D) / flhDC+RBS+IL-12+HlyA+T (ND)/ flhDC+RBS+GFP+HlyA+T (D)/ flhDC+RBS+GFP+HlyA+T (ND)/ SoxS+RBS+GFP+HlyA+T (D)/ SoxS+RBS+GFP+HlyA+T Test (D)/ SoxS+RBS+GFP+HlyA+T (ND) / pConst+RBS+QseB+RBS+QseC+T (D)/ pConst+RBS+QseB+RBS+QseC+T (ND)
Quantification and checking digestion of sequences to ship
Objectives:
- Finishing the assembly of our devices
- Secretion device (containing HlyB, HlyD and TolC in two manners: regulated by flhDC promoter or SoxS promoter)
- Th1-directed device (containing QseC and QseB sensor proteins constitutively expressed and IL-12 whose expression is regulated by flhDC promoter)
- Th2-directed device (containing the SoxR sensor protein constitutively expressed and IL-10 whose expression is regulated by SoxS promoter)
- After finishing the assembly of these parts, we aim to put together the plasmid containing the Secretion system (1.1) and the plasmid containing the sensor-response systems (the one that senses Adrenaline and responds by producing IL-12 (1.2) and the one which senses Nitric Oxide and responds by producing IL-10 (1.3).
Samples:
- Gel 1:
- Ladder / pConst+RBS+SoxR+T C1 (D)/ pConst+RBS+SoxR+T C2 (D)/ pConst+RBS+SoxR+T C3 (D)/ pConst+RBS+SoxR+T C4 (D)/ pConst+RBS+SoxR+T (ND) / SoxS+RBS+IL-10+HlyA+T C1 (D)/ SoxS+RBS+IL-10+HlyA+T C2 (D)/ SoxS+RBS+IL-10+HlyA+T C3 (D)/ SoxS+RBS+IL-10+HlyA+T C4 (D)/ SoxS+RBS+IL-10+HlyA+T (ND)/ flhDC+RBS+IL-12+HlyA+T (D) / flhDC+RBS+IL-12+HlyA+T (ND)/ flhDC+RBS+GFP+HlyA+T (D)/ flhDC+RBS+GFP+HlyA+T (ND)/ SoxS+RBS+GFP+HlyA+T (D)/ SoxS+RBS+GFP+HlyA+T Test (D)/ SoxS+RBS+GFP+HlyA+T (ND) / pConst+RBS+QseB+RBS+QseC+T (D)/ pConst+RBS+QseB+RBS+QseC+T (ND)
- Gel 2:
- Ladder / flhDC+RBS+HlyB+RBS+HlyD+TolC+T (D)/ flhDC+RBS+HlyB+RBS+HlyD+TolC+T (ND)/ SoxS+RBS+ HlyB+RBS+HlyD+TolC+T (D)/ SoxS+RBS+ HlyB+RBS+HlyD+TolC+T (ND)/ pSB1C3 TolC C10 (D) / pSB1C3 TolC C10 (ND)
- Ladder:
- 100-10.000 bp (Fermentas)
- Gel Agarose concentration:
- 1,0%
- Results:
| Construction | Total size (gene+vector) | Linear vector size | Gene size | Result |
|---|---|---|---|---|
| Const-RBS-SoxR-T (C1) | 2753 | 2100 | 653 | smaller |
| Const-RBS-SoxR-T (C2) | 2753 | 2100 | 653 | OK |
| Const-RBS-SoxR-T (C3) | 2753 | 2100 | 653 | No OK |
| Const-RBS-SoxR-T (C4) | 2753 | 2100 | 653 | OK |
| SoxS-RBS-IL10-HlyA-T (C1) | 4170 | 3200 | 970 | smaller |
| SoxS-RBS-IL10-HlyA-T (C2) | 4170 | 3200 | 970 | smaller |
| SoxS-RBS-IL10-HlyA-T (C3) | 4170 | 3200 | 970 | smaller |
| SoxS-RBS-IL10-HlyA-T (C4) | 4170 | 3200 | 970 | smaller |
| flhDC-RBS-IL12-HlyA-T (P2.C1) | 5306 | 3200 | 2106 | OK |
| flhDC-RBS-GFP-HlyA-T (P1.C2) | 3983 | 2800 | 1183 | OK |
| SoxS-RBS-GFP-HlyA-T (C4) | 4360 | 3200 | 1160 | OK |
| SoxS-RBS-GFP-HlyA-T (enzyme test) | 4360 | 3200 | 1160 | OK |
| Const-RBS-QseB-RBS-QseC-T (C1) | 4221 | 2100 | 2121 | strange |
| flhDC-RBS-HlyB-RBS-HlyD-RBS-TolC-T (from competent cells – C2) | 8280 | 2800 | 5480 | OK |
| SoxS-RBS-HlyB-HlyD-TolC-T (from competent cells – P1.C3) | 8257 | 2800 | 5457 | OK |
| pSB1C3-TolC (C10) | 3650 | 2070 | 1580 | No fragment band |

 "
"