Team:TU-Delft/Project/Localization
From 2011.igem.org
(Difference between revisions)
CarstenBlom (Talk | contribs) (Created page with "{{TU-header3}} __NOTOC__ <html> <div style="text-align:center; " > <div style="background-color:#FFFFFF; text-align:left; width:900px; margin:4px auto 10px auto; " id...") |
CarstenBlom (Talk | contribs) (→Localization) |
||
| (2 intermediate revisions not shown) | |||
| Line 15: | Line 15: | ||
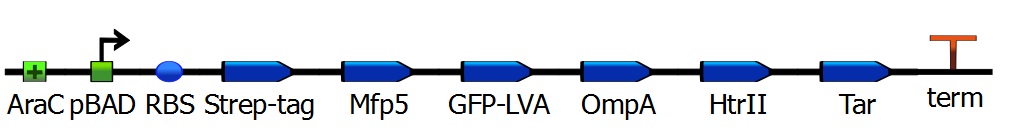
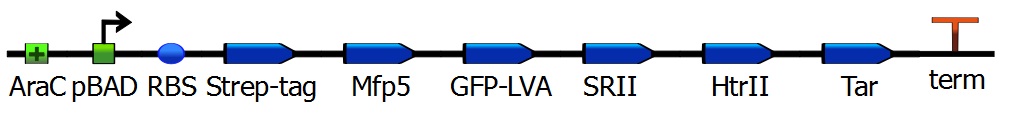
The iGEM team of SDU-Denmark 2010 has produced a phototaxis system.One of the proteins they used was a transmembrane transporter clustered to one specific side of the cell. Denmark achieved this by the use of specific directed proteins, which are involved by the formation of the flagellae, attached to their construct is sensory rhodopsin. Replacing this rhodopsin with our construct, or attaching our Mfp5-GFP to the sensory rhodopsin, enables localization of Mfp5. | The iGEM team of SDU-Denmark 2010 has produced a phototaxis system.One of the proteins they used was a transmembrane transporter clustered to one specific side of the cell. Denmark achieved this by the use of specific directed proteins, which are involved by the formation of the flagellae, attached to their construct is sensory rhodopsin. Replacing this rhodopsin with our construct, or attaching our Mfp5-GFP to the sensory rhodopsin, enables localization of Mfp5. | ||
| - | [[File:Localization_construct_1.jpg]] | + | [[File:Localization_construct_1.jpg|900px]] |
| - | [[File:Localization_construct_2.jpg]] | + | [[File:Localization_construct_2.jpg|900px]] |
{{TU-footer}} | {{TU-footer}} | ||
Latest revision as of 20:29, 21 September 2011








 "
"