Team:TU-Delft/Project/Modelling
From 2011.igem.org
(Difference between revisions)
(→The Regulatory Model) |
K.warringa (Talk | contribs) |
||
| (15 intermediate revisions not shown) | |||
| Line 24: | Line 24: | ||
The intracellular module gives us information about the adhesiveness of Mfp-5 on the membrane which is directly dependent on the inducer (L-arabinose) concentration. It includes all steps in between the inducer (induction of the Mfp-5 expression?) and the amount of Mfp-5 eventually present on the outer membrane, such as for example the influence of the transcription factor, the efficiency of DNA-polymerase and the ribosome concentration in relation to the specific growth rate μ. | The intracellular module gives us information about the adhesiveness of Mfp-5 on the membrane which is directly dependent on the inducer (L-arabinose) concentration. It includes all steps in between the inducer (induction of the Mfp-5 expression?) and the amount of Mfp-5 eventually present on the outer membrane, such as for example the influence of the transcription factor, the efficiency of DNA-polymerase and the ribosome concentration in relation to the specific growth rate μ. | ||
| - | + | https://static.igem.org/mediawiki/2011/8/80/Regulatory_model_include_flux.jpg'' | |
| + | |||
''Extracellular L-arabinose (Ara_out) is transported inside the cell (Ara_in). Here it binds to the transcription factor (TF) which becomes active (TF*) and binds to the promotor (P). The active promotor (P*) is transcribed by DNA-polymerase (pol) and the resulting mRNA is translated by ribosomes (ribo) into intracellular Mfp5-GFP-OmpA (Mfp5_in). This is transported by the Sec transport system to the periplasm. The periplasmic Mfp5-GFP-OmpA (Mfp5_p) folds and inserts itself into the outer membrane (Mfp5_m). When tyrosinase is present tyrosine groups will be hydroxylated to L-DOPA groups resulting in adhesive membrane-bound Mfp5-GFP-OmpA (Mfp5_m*). '' | ''Extracellular L-arabinose (Ara_out) is transported inside the cell (Ara_in). Here it binds to the transcription factor (TF) which becomes active (TF*) and binds to the promotor (P). The active promotor (P*) is transcribed by DNA-polymerase (pol) and the resulting mRNA is translated by ribosomes (ribo) into intracellular Mfp5-GFP-OmpA (Mfp5_in). This is transported by the Sec transport system to the periplasm. The periplasmic Mfp5-GFP-OmpA (Mfp5_p) folds and inserts itself into the outer membrane (Mfp5_m). When tyrosinase is present tyrosine groups will be hydroxylated to L-DOPA groups resulting in adhesive membrane-bound Mfp5-GFP-OmpA (Mfp5_m*). '' | ||
| Line 36: | Line 37: | ||
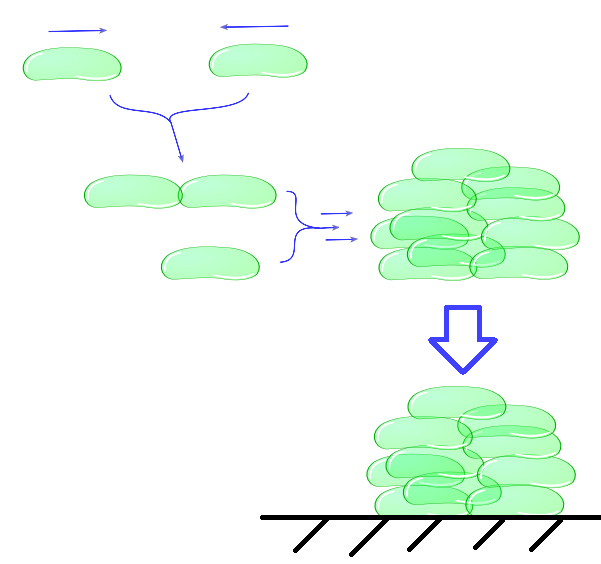
''Individual cells bump into eachother and depending on adhesive strength of the Mfp5 and its concentration they will attach to form a larger cluster. This continuous clustering results into large heavy clusters with a high volume to surface ratio increasing their settling rate.'' | ''Individual cells bump into eachother and depending on adhesive strength of the Mfp5 and its concentration they will attach to form a larger cluster. This continuous clustering results into large heavy clusters with a high volume to surface ratio increasing their settling rate.'' | ||
| - | + | ||
<div style="text-align:center; " > | <div style="text-align:center; " > | ||
<div style="background-color:#FFFFFF; text-align:left; width:900px; margin:4px auto 10px auto; " id="body_content" > | <div style="background-color:#FFFFFF; text-align:left; width:900px; margin:4px auto 10px auto; " id="body_content" > | ||
<Br><Br> | <Br><Br> | ||
| - | |||
| - | |||
== Results == | == Results == | ||
=== The Cluster model === | === The Cluster model === | ||
| + | |||
| + | [https://static.igem.org/mediawiki/2011/7/7b/Cluster_v6-3.pdf For the full script. Please click here!] | ||
| + | |||
| + | |||
| + | [https://static.igem.org/mediawiki/2011/3/32/Bp-6.pdf Bp-6. Please click here!] | ||
| + | |||
| + | |||
| + | [https://static.igem.org/mediawiki/2011/d/d5/Stick-4.pdf Stick-4. Please click here!] | ||
| + | |||
| + | |||
| + | [https://static.igem.org/mediawiki/2011/e/e9/Update_lam_x-3.pdf Lam x-3. Please click here!] | ||
| + | |||
=== The Regulatory Model === | === The Regulatory Model === | ||
| - | + | ====Description==== | |
To perform simulations, a numerical ordinary differential equation solver of MatLab was used (ode45). | To perform simulations, a numerical ordinary differential equation solver of MatLab was used (ode45). | ||
| Line 94: | Line 105: | ||
The arabinose concentration outside the cell was 55 E-5 mol/L, a relatively high concentration, which is expected to cause a high production rate of the protein. (Kleinschmidt et al. (1996)) The maximal produced mfp-5 was produced after 750s. A maximal mfp-5 concentration of 0.038 mmol/L was determined. <br/><br/> | The arabinose concentration outside the cell was 55 E-5 mol/L, a relatively high concentration, which is expected to cause a high production rate of the protein. (Kleinschmidt et al. (1996)) The maximal produced mfp-5 was produced after 750s. A maximal mfp-5 concentration of 0.038 mmol/L was determined. <br/><br/> | ||
| - | + | ====The Matlab Code==== | |
| + | |||
| + | [https://static.igem.org/mediawiki/2011/f/f2/MatlabScript.pdf Please click here!] | ||
| + | |||
Latest revision as of 21:46, 3 February 2012








 "
"