Team:TU-Delft/Project/Modelling
From 2011.igem.org
(Difference between revisions)
CarstenBlom (Talk | contribs) (→Modelling) |
CarstenBlom (Talk | contribs) (→Modelling) |
||
| Line 25: | Line 25: | ||
[[File:Mechanical_model_regulatory_level.jpg|400px]] | [[File:Mechanical_model_regulatory_level.jpg|400px]] | ||
| + | |||
| + | |||
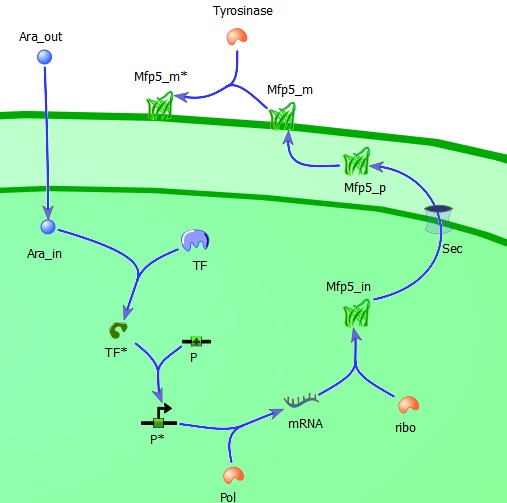
''Extracellular L-arabinose (Ara_out) is transported inside the cell (Ara_in). Here it binds to the transcription factor (TF) which becomes active (TF*) and binds to the promotor (P). The active promotor (P*) is transcribed by DNA-polymerase (pol) and the resulting mRNA is translated by ribosomes (ribo) into intracellular Mfp5-GFP-OmpA (Mfp5_in). This is transported by the Sec transport system to the periplasm. The periplasmic Mfp5-GFP-OmpA (Mfp5_p) folds and inserts itself into the outer membrane (Mfp5_m). When tyrosinase is present tyrosine groups will be hydroxylated to L-DOPA groups resulting in adhesive membrane-bound Mfp5-GFP-OmpA (Mfp5_m*). '' | ''Extracellular L-arabinose (Ara_out) is transported inside the cell (Ara_in). Here it binds to the transcription factor (TF) which becomes active (TF*) and binds to the promotor (P). The active promotor (P*) is transcribed by DNA-polymerase (pol) and the resulting mRNA is translated by ribosomes (ribo) into intracellular Mfp5-GFP-OmpA (Mfp5_in). This is transported by the Sec transport system to the periplasm. The periplasmic Mfp5-GFP-OmpA (Mfp5_p) folds and inserts itself into the outer membrane (Mfp5_m). When tyrosinase is present tyrosine groups will be hydroxylated to L-DOPA groups resulting in adhesive membrane-bound Mfp5-GFP-OmpA (Mfp5_m*). '' | ||
| Line 33: | Line 35: | ||
[[File:Mechanical_model_intercellular_module.png|400px]] | [[File:Mechanical_model_intercellular_module.png|400px]] | ||
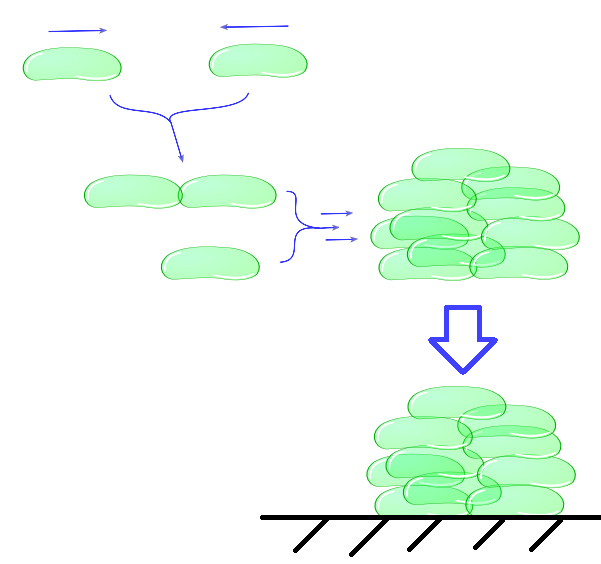
| - | + | ''Individual cells will bump into eachother and when adhesiveness is strong enough will attach to form a larger cluster. This continuous clustering results into large heavy clusters with a high volume to surface ratio increasing their settling rate.'' | |
Revision as of 20:48, 21 September 2011








 "
"