Team:Cambridge/Project
From 2011.igem.org
(→Objective One - Express reflectin in E. coli) |
(→Objective One - Express reflectin in E. coli) |
||
| Line 47: | Line 47: | ||
=Objective One - Express reflectin in E. coli= | =Objective One - Express reflectin in E. coli= | ||
| - | Our first objective was to try to express reflectin, in any shape or form, in E. coli. Ideally, we'd be able to express reflectin in the bacteria at a range of levels using a single construct, so we can both overexpress (for ''in vitro'' studies) or underexpress (to promote ''in vivo'' folding) straightforwardly. In order to do this, we had to create an expression plasmid for reflectin with which we could transform the bacterial cells. Hence, we had to isolate a reflectin gene sequence, choose a suitable promotor and join them on an appropriate backbone in order to engineer what we wanted. | + | Our first objective was to try to express reflectin, in any shape or form, in E. coli. Ideally, we'd be able to express reflectin in the bacteria at a range of levels using a single construct, so we can both overexpress (for ''in vitro'' studies) or underexpress (to promote ''in vivo'' folding) straightforwardly. In order to do this, we had to create an expression plasmid for reflectin with which we could transform the bacterial cells. Hence, we had to isolate a reflectin gene sequence, choose a suitable promotor and join them on an appropriate backbone in order to engineer what we wanted. These steps are outlined below. |
[[File:camflow_1.jpg | 600px | Objective One]] | [[File:camflow_1.jpg | 600px | Objective One]] | ||
| + | ==Isolating the reflectin gene== | ||
| + | |||
| + | ==Selecting the promotor== | ||
| + | |||
| + | ==Selecting the plasmid== | ||
| + | |||
| + | ==Join | ||
=='''References'''== | =='''References'''== | ||
Revision as of 11:17, 10 August 2011
Contents |
Background to our project
How to Disappear Completely - Reflectins in cephalopods
The beautiful optical properties which reflectins make possible are used for a multitude of purposed within nature. Manipulation of reflectance allows squids to communicate through polarised light whilst altering the refractive index to match that of the water column allows cephalopods to hide from their predators, the very replication of a living invisibility cloak.
Reflectin was first identified as such in the Hawaiian bobtail squid [http://en.wikipedia.org/wiki/Euprymna_scolopes Euprymna scolopes] as the protein responsible for a reflective layer in the "light organ". This layer reflects light emitted by symbiotic bacteria to be reflected downwards away from the squid for camouflage, like a [http://en.wikipedia.org/wiki/Headlamp#Reflector_lamps car headlamp].
Whereas the effect is static in E.scolopes, the Longfin inshore squid [http://en.wikipedia.org/wiki/Longfin_Inshore_Squid Loligo pealei] demonstrates dynamic iridescence which are controlled by the nervous system[5]. The neural-signals switches a [http://en.wikipedia.org/wiki/Kinase protein kinase], an enzyme which adds negatively charged phosphate groups to the positively charged protein. Within the squid reflectin is structured hierarchially, the structure of the reflectin protein itself, the complex platelets which form and the final multi-layer 'Bragg stack' of platelets contained within reflective cells called iridophores[3]. The negatively charged phosphate groups alters the spacing between the protein layers and therefore the colour by changing the attraction/repulsion between the reflectin platelets.(Read more about structural colour)
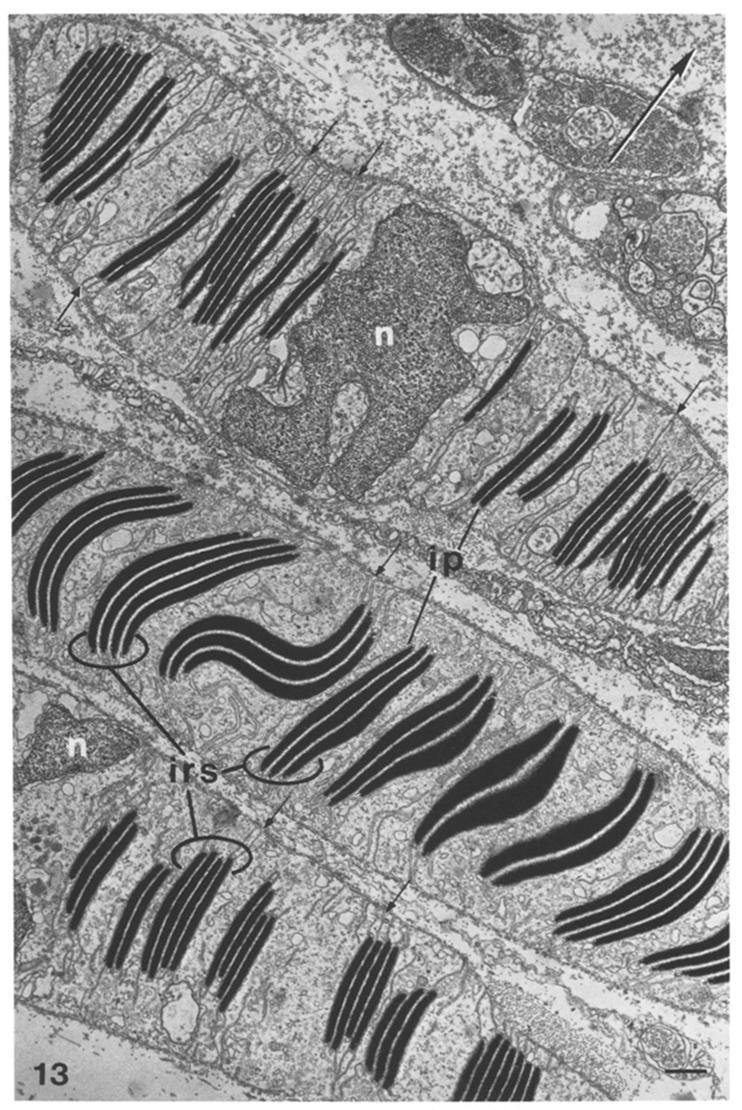
EM of reflectin ultrastructure (dark bands) in cells from Loligo opalescens[6] |
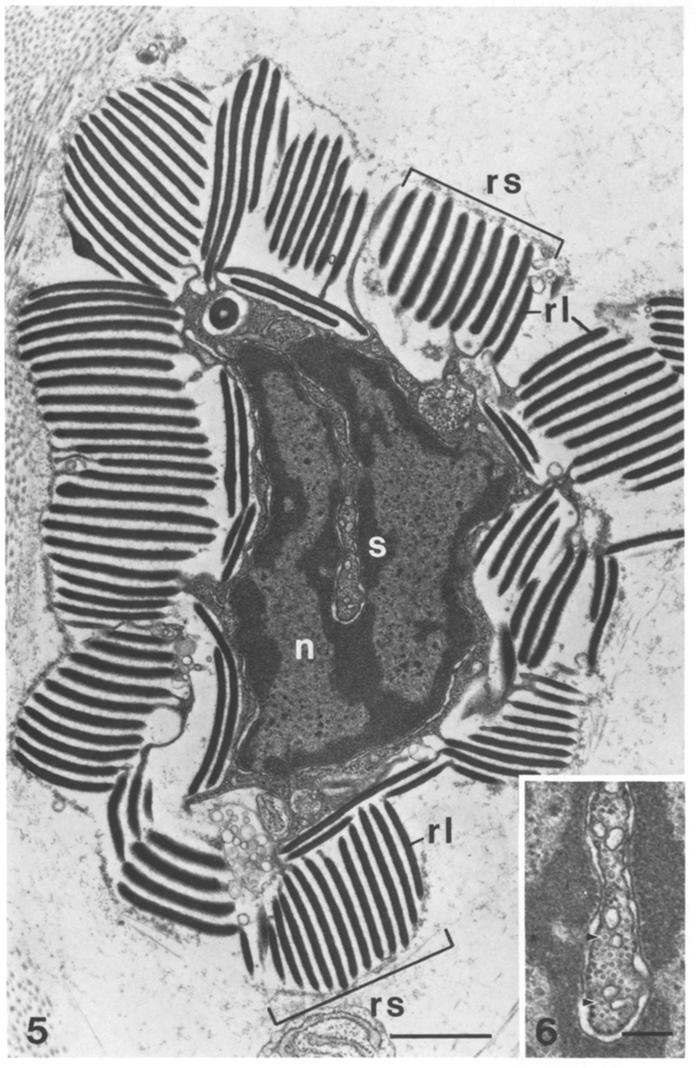
EM of reflectin ultrastructure (dark bands) in reflector cells from Octopus dofleini[6] |
Iridophores are not merely confined in cephalopods but can also be found in other members of mollusc phylum e.g. giant clams[4]where they are responsible for the stunning iridescent colours of the mantle, and they may play a role in protecting against harmful UV and maximising capture of sunlight for the photosynthetic symbionts.
Light and interference - The physics behind structural colour
Structural colour is a common occurrence in nature - butterfly wings, fish scales and even [http://en.wikipedia.org/wiki/Tapetum_lucidum cats's eyes] contain light reflecting components rather than pigments. It occurs due to the phenomenon of [http://en.wikipedia.org/wiki/Thin-film_interference thin film interference].
The difference in [http://en.wikipedia.org/wiki/Refractive_index refractive index] between the alternating layers leads to reflection (bouncing off) and transmission of light. This process is repeated at each layer. When the two reflected light waves meet, they sum together but are no longer in sync (due to differences in the optical paths they have taken) leading to the phenomenon known as interference. Some wavelengths will destructively interfere more than others. The result is that not all colours are reflected with the same intensity, creating rainbow-like effects like those of oil droplets on the surface of water or the surface of shimmering soap bubbles.The observed colour changes with viewing angle, angle of incidence of light and alterations in the spacing of the layers or refractive indices which alters the mechanism of interference. In fact, if you use layers of materials with matching refractive indices, total transparency and hence invisibility of the material can be achieved.
This principle of interference is believed to be the primary mechanism by which colour is controlled in the skin of squid. Experimental observations of changes in the thickness and spacing between reflectin layers in-vivo has been observed following post translational modification.
Reflectins as Novel Polymers
Reflectins unique properties partly stem from their unique amino acid composition; residues normally common in proteins (alanine, isoleucine, leucine and lysine) are nowhere to be found in any reflectins identified thus far. Meanwhile typically rare residues (arginine, methionine, tryptophan and tyrosine) make up almost 57% of the protein! The family of reflectin proteins also share a repeated domain which could also possess some unique optical characteristics.
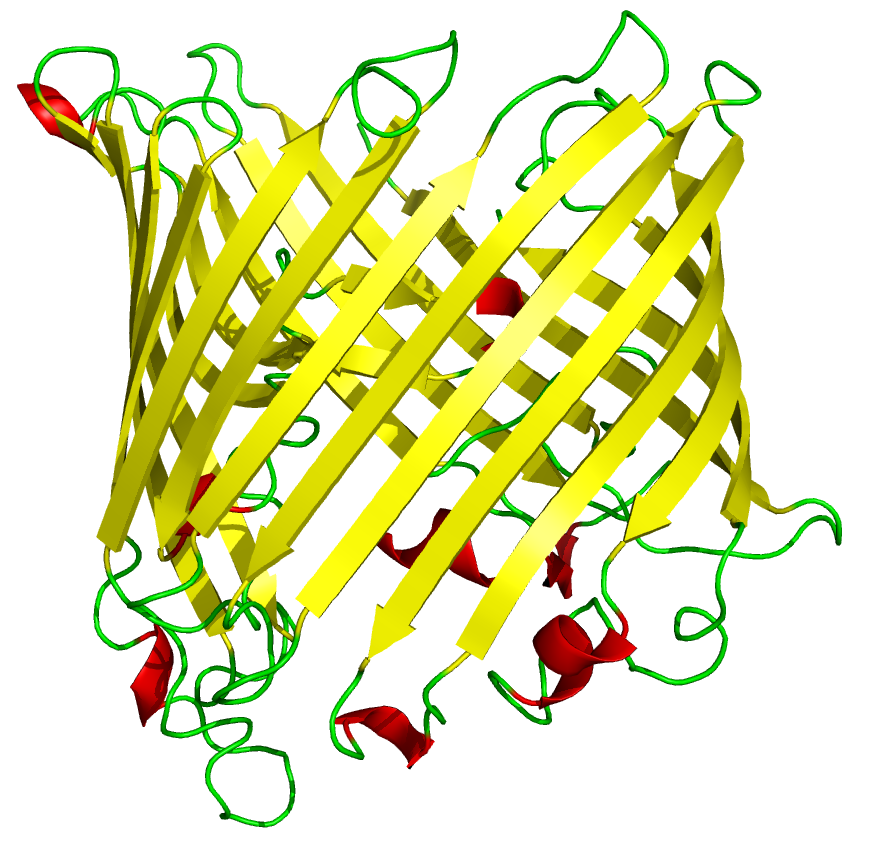
The 3D protein structure of reflectin has not yet been documented. It may be natively unstructured, Weiss et al hypothesised that it may form a [http://en.wikipedia.org/wiki/Beta_barrel beta barrel] like structure when interacting with membranes, as recombinant reflectin-like proteins associates strongly with artificial membrane structures after cell-free expression.
Kramer et al and Tao and DeMartini et al have demonstrated the remarkable capability of reflectin proteins to self assemble in vitro to create complex structures. Recombinant reflectin, refolded in vitro, can be carefully spread along a silicon slide to make thin films with intense structural colours which showed iridescence. Measurement of the refractive index revealed reflectin possessing the highest refractive index of any natural known protein. In addition Kramer et al demonstrated the ability of reflectin to form a diffraction grating when ionic solvents used to dissolve it, were diffused away in a water bath.

Objective One - Express reflectin in E. coli
Our first objective was to try to express reflectin, in any shape or form, in E. coli. Ideally, we'd be able to express reflectin in the bacteria at a range of levels using a single construct, so we can both overexpress (for in vitro studies) or underexpress (to promote in vivo folding) straightforwardly. In order to do this, we had to create an expression plasmid for reflectin with which we could transform the bacterial cells. Hence, we had to isolate a reflectin gene sequence, choose a suitable promotor and join them on an appropriate backbone in order to engineer what we wanted. These steps are outlined below.
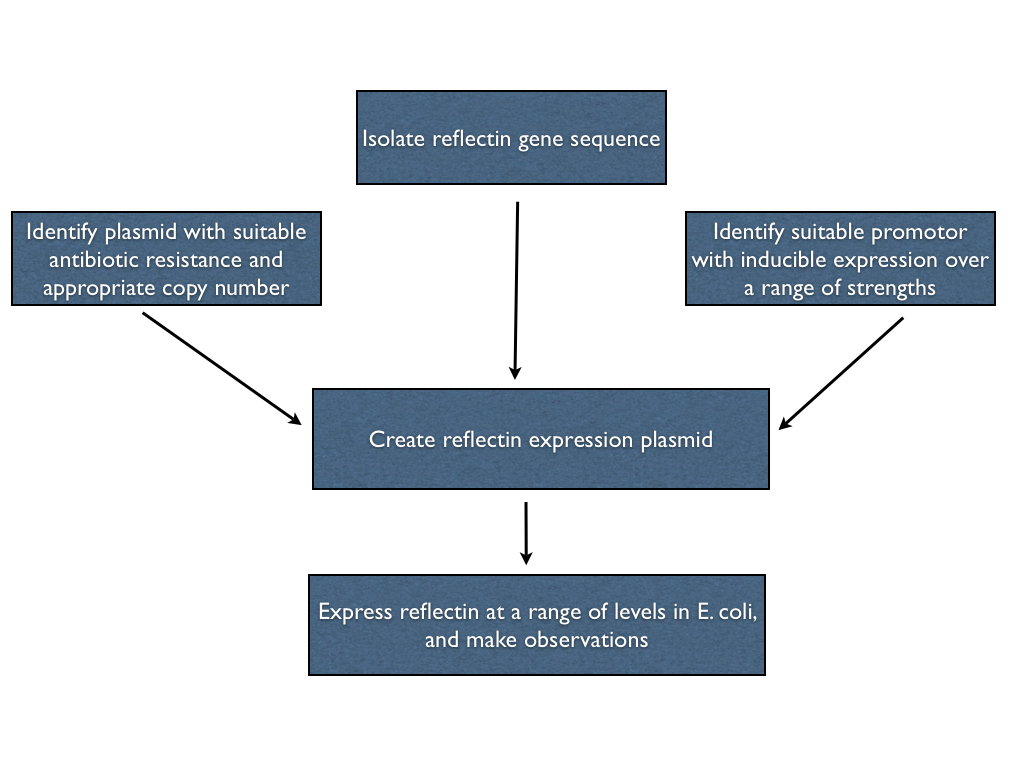
Isolating the reflectin gene
Selecting the promotor
Selecting the plasmid
==Join
References
[http://www.nature.com/nmat/journal/v6/n7/abs/nmat1930.html] Kramer et al. The self-organizing properties of squid reflectin protein Nature Materials 533-538 VOL6 JULY 2007 [http://www.sciencemag.org/content/303/5655/235.short] Crookes et al. Reflectins: The Unusual Proteins of Squid Reflective Tissues SCIENCE 235-238 VOL303 9 JANUARY 2004 [http://www.sciencedirect.com/science/article/pii/S0142961209011442] Morse et al. The role of protein assembly in dynamically tunable bio-optical tissues Biomaterials 793-801 VOL31 FEBRUARY 2010 [http://www.publish.csiro.au/paper/ZO9920319.html] Griffiths et al. Iridophores in the mantle of giant clams. Australian Journal of Zoology (1992) Volume: 40, Issue: 3 Pages: 319-326 [http://www.ncbi.nlm.nih.gov/pubmed/19776150] Izumi et al. Changes in reflectin protein phosphorylation are associated with dynamic iridescence in squid. J. R. Soc. Interface 6 March 2010 vol. 7 no. 44 549-560 [http://www.springerlink.com/content/bba14b73ad35f495/]Brocco et al. Reflector cells in the skin of Octopus dofleini Cell and Tissue Research, Volume 205, Number 2, 167-186, 1980
 "
"