Team:Cambridge/Project
From 2011.igem.org
(→Isolating the reflectin gene) |
(→Achievements) |
||
| (43 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | ||
| + | Bact<b>iridescence</b> was based around the properties of [[Team:Cambridge/Project/Background | reflectin]], a squid protein with the highest refractive index of any known proteinaceous substance. In squid this protein forms complex platelets which act as [http://en.wikipedia.org/wiki/Bragg_reflector Bragg reflectors] to provide camouflage. | ||
| - | + | ===Project Goals=== | |
| - | + | We aimed to [[Team:Cambridge/Project/In_Vivo | express reflectin in ''E. coli'']] and to investigate its optical properties in order to build the groundwork for the manipulation of living structural colour. We also looked at the [[Team:Cambridge/Project/In_Vitro | over-expression of reflectin in ''E. coli'']], in order to obtain relatively pure samples of the protein for making thin films. | |
| - | + | Much of our work (particularly the in vivo work) simply hadn't been tried before, so, while we had high hopes, we could not be sure as to what would happen. | |
| - | + | ||
| - | + | ||
| - | = | + | ==Achievements== |
| - | + | In one short summer the [[Team:Cambridge/Team | 2011 Cambridge team]] has produced a set of [[Team:Cambridge/Parts | BioBrick parts]] to allow future researchers to explore synthetic biology applications for structural colour. | |
| - | [[ | + | |
| - | == | + | ===[[Team:Cambridge/Project/In_Vivo | In Vivo]]=== |
| - | + | Working with living cells we have; | |
| + | *[[Team:Cambridge/Project/Microscopy | Imaged squid tissue using novel techniques]] to explore the in vivo properties of reflectins. | ||
| + | *Succesfully produced reflectins in ''E. coli''. | ||
| + | *Characterised best practices for in vivo reflectin production. | ||
| - | == | + | ===[[Team:Cambridge/Project/In_Vitro | In Vitro]]=== |
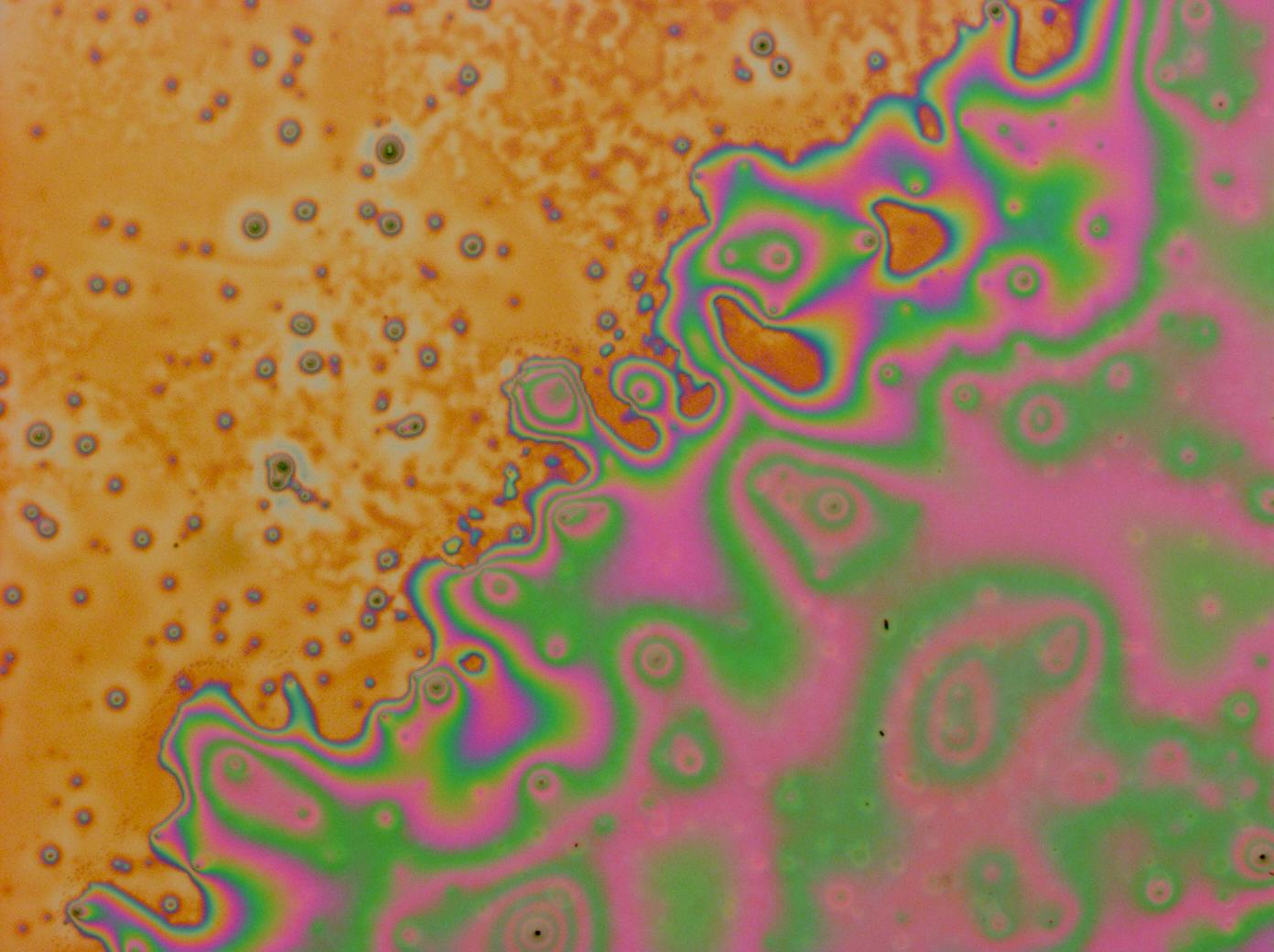
| + | [[File:Cam_Multilayer_drop_1.jpg | right | thumb | 150px | A multilayer thin film]] | ||
| + | By engineering ''E. coli'' to overexpress reflectins we have; | ||
| + | *[[Team:Cambridge/Experiments/Protein_Purification | Purified reflectin]] and documented best practice for high purity yields. | ||
| + | *Made [[Team:Cambridge/Project/Microscopy#Reflectin_Thin_Films | thin films]] which show structural colours. | ||
| + | *Demonstrated the rapid colour changes possible with reflectin. | ||
| + | **Videos of our thin films are available on [http://www.youtube.com/user/cambridgeigem2011 youtube]. | ||
| - | == | + | ===[[Team:Cambridge/Project/Gibthon | Software]]=== |
| + | [[File:Gibthon2.0beta.png | left | thumb | 100px | The Gibthon logo]] | ||
| + | We contributed to [http://www.gibthon.org/ Gibthon], an open-source collection of web-based tools for construct design, fully compatible with both BioBrick standards and newer assembly techniques. | ||
| + | *Greatly improved import and display of fragments (including support for [http://partsregistry.org/Main_Page partsregistry.org]). | ||
| + | *Added tools to allow management of uploaded parts. | ||
| - | = | + | <html><div style='clear:both'></div></html> |
| - | + | ||
| - | == | + | ==[[Team:Cambridge/Project/Future | Future work]]== |
| - | + | ||
| + | By creating the first BioBrick parts for production of structural colour, we hope to facilitate further research. Although time did not allow us to explore the full potential of our project, we have some ideas for what could be done next. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | ||
Latest revision as of 02:52, 22 September 2011
Contents |
Project Goals
We aimed to express reflectin in E. coli and to investigate its optical properties in order to build the groundwork for the manipulation of living structural colour. We also looked at the over-expression of reflectin in E. coli, in order to obtain relatively pure samples of the protein for making thin films.
Much of our work (particularly the in vivo work) simply hadn't been tried before, so, while we had high hopes, we could not be sure as to what would happen.
Achievements
In one short summer the 2011 Cambridge team has produced a set of BioBrick parts to allow future researchers to explore synthetic biology applications for structural colour.
In Vivo
Working with living cells we have;
- Imaged squid tissue using novel techniques to explore the in vivo properties of reflectins.
- Succesfully produced reflectins in E. coli.
- Characterised best practices for in vivo reflectin production.
In Vitro
By engineering E. coli to overexpress reflectins we have;
- Purified reflectin and documented best practice for high purity yields.
- Made thin films which show structural colours.
- Demonstrated the rapid colour changes possible with reflectin.
- Videos of our thin films are available on [http://www.youtube.com/user/cambridgeigem2011 youtube].
Software
We contributed to [http://www.gibthon.org/ Gibthon], an open-source collection of web-based tools for construct design, fully compatible with both BioBrick standards and newer assembly techniques.
- Greatly improved import and display of fragments (including support for [http://partsregistry.org/Main_Page partsregistry.org]).
- Added tools to allow management of uploaded parts.
Future work
By creating the first BioBrick parts for production of structural colour, we hope to facilitate further research. Although time did not allow us to explore the full potential of our project, we have some ideas for what could be done next.
 "
"