Team:Cambridge/Protocols/PCR
From 2011.igem.org
Contents |
Polymerase Chain Reaction (PCR)
The goal of PCR is to amplify a section of DNA of interest for DNA analysis (e.g. gene insertion, sequencing, etc). The amplification rate is exponential.
Theory
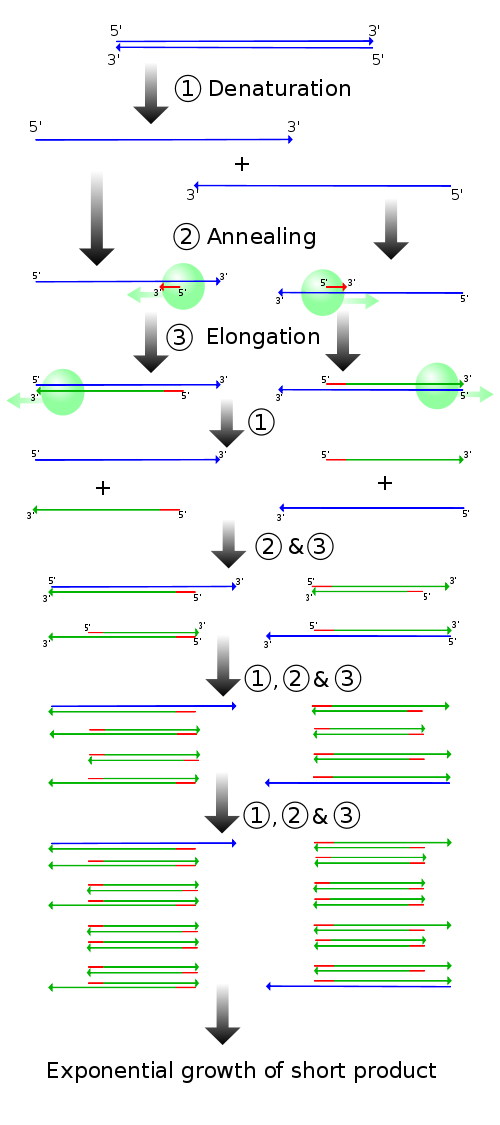
(1) Denaturing at 94–96 °C
(2) Annealing at ~65 °C
(3) Elongation at 72 °C Four cycles are shown here. The blue lines represent the DNA template to which primers ( red arrows ) anneal that are extended by the DNA polymerase ( light green circles ), to give shorter DNA products ( green lines ), which themselves are used as templates as PCR progresses. Credit, WikiMediaThe reaction works by the annealing of primers to single stranded DNA, which are extended by DNA polymerase. By repeating the process, the section of DNA between the primers is amplified.
TODO: More Here!
Practice
The PCR reaction with Phusion Hot Start II DNA Polymerase contains the following reagents:
| Name | 50 μl reaction | 20 μl reaction | Final concentration |
|---|---|---|---|
| De-Ionized Water | add to 50 μl | add to 20 μl | |
| 5x Phusion Buffer | 10 μl | 4 μl | 1x |
| 10mM dNTPs | 1 μl | 0.4 μl | 200μM each |
| Forward Primer | x μl | x μl | 0.5 μM |
| Reverse Primer | x μl | x μl | 0.5 μM |
| Template DNA | x μl | x μl | |
| Phusion Polymerase | 0.5 μl | 0.2 μl | 0.02 U/μl |
Tip: The recommended buffer is 5x Phusion HF Buffer, used as a default buffer for high fidelity amplification.
Tip: When you receive primers synthesized by a biotechnological company
- centrifuge tubes prior to opening to prevent loss of pelleted oligonucleotides
- add the amount of water specified in the table provided with primers to obtain 100 μM solution
- prepare a 4-fold dilution of a portion of the primer solution
Only the primers and the DNA template are specific to the reaction. The remaining reagents can be made as a 'Master Mix' in order to reduce the need to repeatedly pipette small volumes which amplify experimental error.
As with all enzyme-containing reagents, the master mix should be kept in the freezer and small aliquots thawed when required.
The remainder of the reaction is handled automatically by a PCR machine. It is common practice to perform a gel electrophoresis to extract the correct DNA after PCR.
An example of conditions we used for a PCR reaction using Phusion polymerase are as follows:
| Cycle | No. cycles | Step | Temp (C) | Time (s) |
| 1 | 1 | 1 | 98 | 30 |
| 2 | 30 | 1 | 98 | 10 |
| 2 | 55 | 30 | ||
| 3 | 72 | 2:30 | ||
| 3 | 1 | 1 | 72 | 5 |
| 4 | 50 | 1 | 4 | 99.59 (end) |
Safety
No bacteria are used during the reaction there is therefore little or no biological hazard. Nevertheless, it is important to observe correct laboratory procedure and wear appropriate clothing and gloves. PCR occurs at high temperature, and this may present a risk, depending on the PCR machine employed.
 "
"
