Team:Cambridge/Experiments/Initial Exercise Group One
From 2011.igem.org
Contents |
Initial Exercise: Katy, Heather, Marta and Veronica
The aim of the project was to produce interesting patterns of fluorescence in a colony of Bacillus Subtilis bacteria by fusion of a single gene of interest in the bacillus ge=nome to GFP using Gibson Assembly. These patterns would need to be clearly observable using a confocal microscope. After some research, our group decided to fuse GFP to the DivIVA gene.
Role of DivIVA
DivIVA is believed to control septum positioning in Bacillus Subtilis. This was investigated (in [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1997.3811764.x/abstract this] study by Edwards and Errington) by considering the phenotype of mutants produced when certain bases in the gene were altered.
Mutants showed:
- inhibition of cell division initiation*
- abnormally placed septum*
- some small anucleate cells*
We also hoped that this fusion would have a high chance of success, since a successful DivIVA-GFP fusion had already been documented, indicating that fluorescence was constitutively expressed at all sites of cell division, new and old, in the bacteria. We therefore know what characteristics to expect and so we should be able to determine whether or not the experiment was successful almost immediately under the microscope.
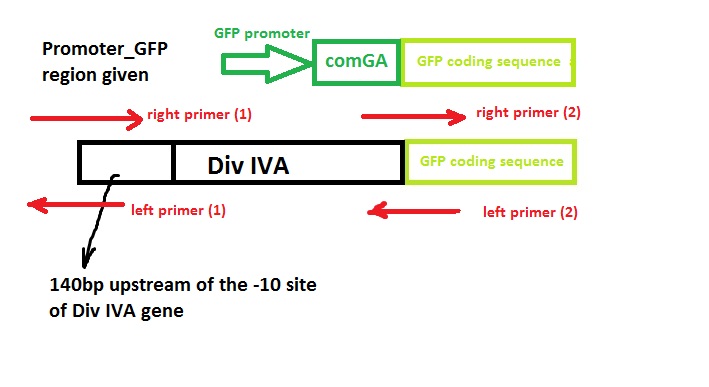
Construct Design
We wish to:
- keep the original promotor for DivIVA*
- remove the GFP promotor sequence*
- remove the DivivA stop codon so that GFP is translated together with the DivIVA protein*
- Remove the ComGA stabiliser sequence pre-existing on the plasmid vector*
‘Linker’ code was also used by Edwards and Errington - however, we suspect that this is related to the restriction enzyme assembly process they used and hence we didn’t include it in our design.
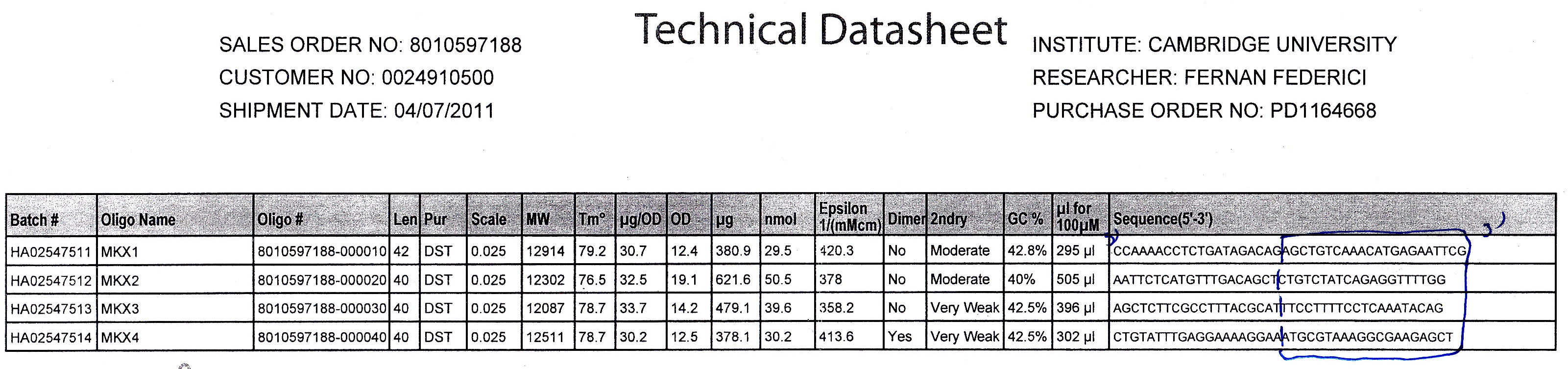
Primer Design
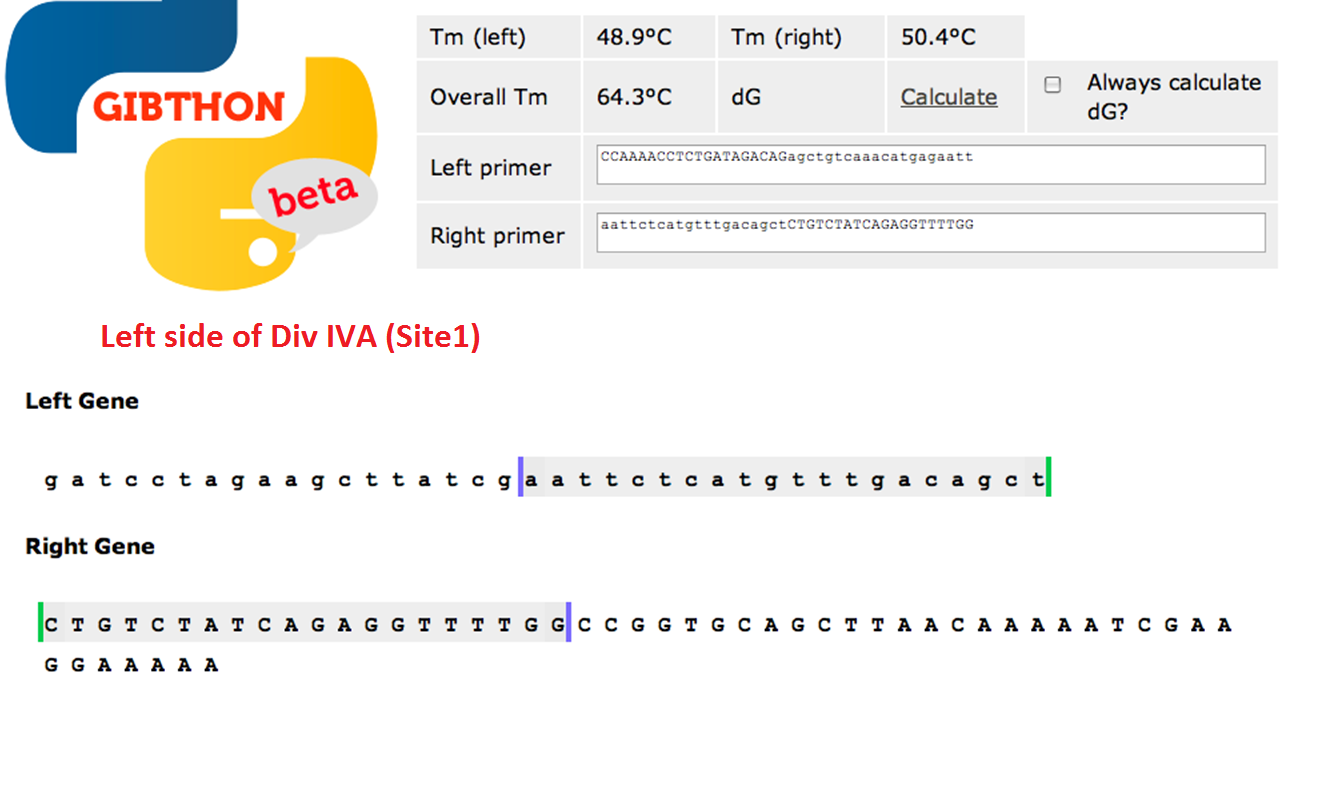
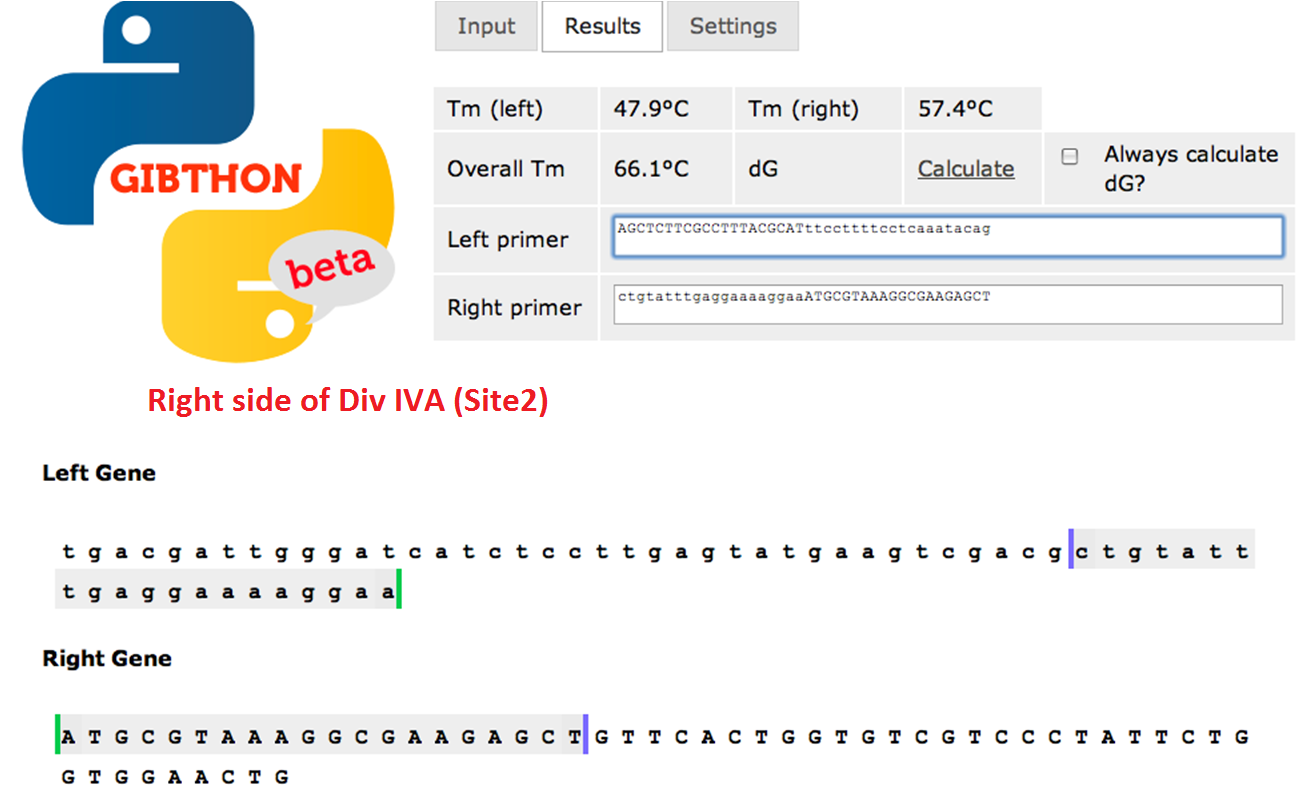
We used the Gibthon Construct Designer developed by the 2010 iGEM Cambridge team to aid us when designing our primers; we did however alter one of the primers slightly in order to achieve similar melting temperatures for primers that would operate on the same DNA fragment.
Left side of the Div IVA coding sequence (site 1)
Right side of the Div IVA coding sequence (site 2)
Predicted results
- Bacillus bacteria constitutively express fluorescence
- Fluorescence seen at both old and new sites of cell division
Method
- 20bp primers with 20bp tails were designed to perform PCR to amplify our chosen DNA sequences and Gibson Assembly to fuse the SrfA promoter with a plasmid containing GFP.
- 100 μM stock solutions of the primers were made and subsequently diluted four-fold to make 25μM 'working solutions'
- Particular care was taken in preparing the inital stock as these affect all future dilutions and were 'contingency' solutions in case of mistake.
- The genomic DNA of B. subtilis and the vector plasmid containing GFP were provided.
- The vector plasmid is to be cloned in two fragments (otherwise the sequence would be too long for effective PCR), so two additional primers were required. These were provided for us in the appropriate concentration.
Need to check primer names
Our three PCR reactions consisted of:
- 1.
- 1μl primer JMF2
- 1μl primer JMF3
- 1μl B. subtilis genomic DNA
- 2.
- 1μl primer JMF1
- 1μl primer A Forward (provided)
- 1μl vector DNA
- 3
- 1μl primer JMF4
- 1μl primer B reverse (provided)
- 1μl vector DNA
For setup 1, to each tube we also added:
- 9.5μl of water
- 12.5μl Master mix (SyBR Green and Rox, Hotstart Taq polymerase, dNTPs and dyes)
for a total volume 25μl
The complete tubes were run in a real-time PCR machine (so that we would be able to observe the progress of our very first PCR reaction in real time) for 30 cycles with a 2 minute extension time and a primer annealing temperature of 50ºc.
In setup 2 we used the same primers and templates as above, but we made a new master mix. This included phusion, a much more processive polymerase. The total volume of master mix was 22ul, which was comprised of:
5ul buffer 0.5ul dNTP 0.25ul phusion polymerase 16.25ul H2O
Technical Data
We used Finnzymes melting temperature calculator to work out the melting temperature of the primer part (not the tail) of our Gibson Assembly oligos (this was the 3' 20bp of each oligo) these values in ºC are shown below Needs to be changed JMF1 - 56.91 JMF2 - 68.8 JMF3 - 50.48 JMF4 - 53.09
Results for Experiment 1
- Reaction 1 and 3 were amplified in the PCR however reaction 2 was not
- It is suspected this was a result of a compromise run of the PCR where:
- Annealing temperature set at an arbitrary 50ºc to produce annealling for every group
- Primer extension time was arbitrarily set at 3 mins
- Results from all groups indicated the PCR failed for the longest fwdA fragment. We ascribe this to the use of Taq polymerase instead of Phusion polymerase, since Phusion produces much higher fidelity.
- It is intended to carry out the PCR again using Phusion polymerase.
Results for Experiment 2
 "
"