|
|
| (93 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} |
| | + | Bact<b>iridescence</b> was based around the properties of [[Team:Cambridge/Project/Background | reflectin]], a squid protein with the highest refractive index of any known proteinaceous substance. In squid this protein forms complex platelets which act as [http://en.wikipedia.org/wiki/Bragg_reflector Bragg reflectors] to provide camouflage. |
| | | | |
| - | {|align="justify"
| + | ===Project Goals=== |
| - | |The Cambridge 2011 iGEM team is made up of nine undergraduates from diverse disciplines. We aim to achieve something remarkable using synthetic biology within the short timeslot alloted for our work. If you would like to sponsor Team Bactiridescence please browse our [https://static.igem.org/mediawiki/2011/c/cb/IGEM_brochure_Cambridge_Team_2011_small_version.pdf brochure ].
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |
| + | |
| - | '''Bact''iridescence''''' is a project based around the unique properties of reflectin, a squid protein with the highest refractive index of any known proteinaceous substance. In squid this protein forms complex platelets which act as [http://en.wikipedia.org/wiki/Distributed_Bragg_reflector Bragg reflectors] to provide camouflage. We aim to express reflectin in E. coli and optimise the optical properties, building the groundwork for the manipulation of living structural colour.
| + | |
| | | | |
| - | |[[Image:Cambridge_team.png|right|frame|Your team picture]]
| + | We aimed to [[Team:Cambridge/Project/In_Vivo | express reflectin in ''E. coli'']] and to investigate its optical properties in order to build the groundwork for the manipulation of living structural colour. We also looked at the [[Team:Cambridge/Project/In_Vitro | over-expression of reflectin in ''E. coli'']], in order to obtain relatively pure samples of the protein for making thin films. |
| - | |-
| + | |
| - | |
| + | |
| - | |align="center"|[[Team:Cambridge | Bactiridescence]]
| + | |
| - | |}
| + | |
| | | | |
| | + | Much of our work (particularly the in vivo work) simply hadn't been tried before, so, while we had high hopes, we could not be sure as to what would happen. |
| | | | |
| - | ==Background== | + | ==Achievements== |
| | + | In one short summer the [[Team:Cambridge/Team | 2011 Cambridge team]] has produced a set of [[Team:Cambridge/Parts | BioBrick parts]] to allow future researchers to explore synthetic biology applications for structural colour. |
| | | | |
| | + | ===[[Team:Cambridge/Project/In_Vivo | In Vivo]]=== |
| | + | Working with living cells we have; |
| | + | *[[Team:Cambridge/Project/Microscopy | Imaged squid tissue using novel techniques]] to explore the in vivo properties of reflectins. |
| | + | *Succesfully produced reflectins in ''E. coli''. |
| | + | *Characterised best practices for in vivo reflectin production. |
| | | | |
| - | ==How to disappear completely (or stand out in a crowd) - ''Reflectins in cephalopods''== | + | ===[[Team:Cambridge/Project/In_Vitro | In Vitro]]=== |
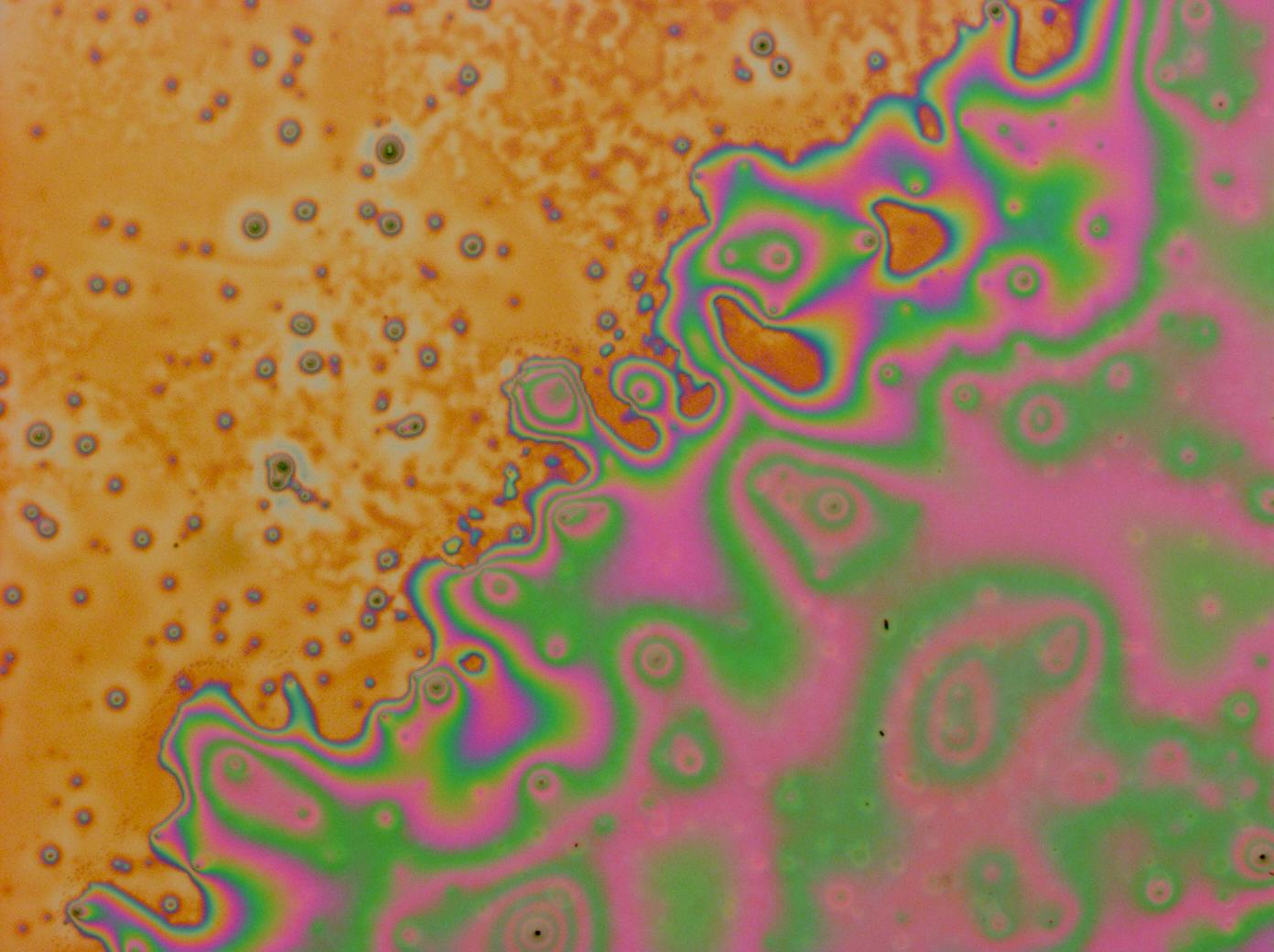
| | + | [[File:Cam_Multilayer_drop_1.jpg | right | thumb | 150px | A multilayer thin film]] |
| | + | By engineering ''E. coli'' to overexpress reflectins we have; |
| | + | *[[Team:Cambridge/Experiments/Protein_Purification | Purified reflectin]] and documented best practice for high purity yields. |
| | + | *Made [[Team:Cambridge/Project/Microscopy#Reflectin_Thin_Films | thin films]] which show structural colours. |
| | + | *Demonstrated the rapid colour changes possible with reflectin. |
| | + | **Videos of our thin films are available on [http://www.youtube.com/user/cambridgeigem2011 youtube]. |
| | | | |
| | + | ===[[Team:Cambridge/Project/Gibthon | Software]]=== |
| | + | [[File:Gibthon2.0beta.png | left | thumb | 100px | The Gibthon logo]] |
| | + | We contributed to [http://www.gibthon.org/ Gibthon], an open-source collection of web-based tools for construct design, fully compatible with both BioBrick standards and newer assembly techniques. |
| | + | *Greatly improved import and display of fragments (including support for [http://partsregistry.org/Main_Page partsregistry.org]). |
| | + | *Added tools to allow management of uploaded parts. |
| | | | |
| - | The beautiful optical properties which reflectin makes possible are used for different purposes in nature. Manipulation of reflectance may allow squid to communicate through polarised light. Altering refractive index to match that of the water column allows cephalopods to hide from their predators - a living invisibility cloak.
| + | <html><div style='clear:both'></div></html> |
| | | | |
| - | Reflectin was first identified as such in the Hawaiian bobtail squid [http://en.wikipedia.org/wiki/Euprymna_scolopes|''Euprymna scolopes''] as the protein responsible for a reflective layer in the "light organ". This allows light emitted by symbiotic bacteria to be reflected downwards away from the squid, like a [http://en.wikipedia.org/wiki/Headlamp#Reflector_lamps|car headlamp].
| + | ==[[Team:Cambridge/Project/Future | Future work]]== |
| | | | |
| - | The Longfin inshore squid [http://en.wikipedia.org/wiki/Longfin_Inshore_Squid|''Loligo pealei''] shows dynamic iridescence controlled through signals from the nervous system which turn on a [http://en.wikipedia.org/wiki/Kinase|protein kinase], an enzyme which adds negatively charged phosphate groups to the positively charged protein. This alters the attraction/repulsion between the platelet arrangement of reflectins, changing the spacing of the iridophore layers and therefore the colour of light reflected. (Read more about structural colour)
| + | By creating the first BioBrick parts for production of structural colour, we hope to facilitate further research. Although time did not allow us to explore the full potential of our project, we have some ideas for what could be done next. |
| | | | |
| - | Squid skin contains multiple specialised cell types designed to allow highly controlled changes of colour and reflectance to optimise camouflage. The beautiful optical properties in light reflecting tissues occur due to a hierarchy of structural arrangements - the structure of reflectin protein itself, the complex platelets which the protein forms, and the shape and layering of reflective cells. A thin tissue layer made up of iridocytes close to the surface of squid skin is made up of ~40% reflectin (out of the total protein content). <sup>[[#Crookes|2]]</sup> Within iridocytes, reflectins self assemble to form membrane associated platelets. The changes in refractive index as light moves through the layers of reflectin and cytoplasm forms a natural '[[#Bragg|Bragg reflector]]'<sup>[[#Morse|[3]]]</sup>.
| |
| - |
| |
| - | Iridophores which appear similar in structure to those found in cephalopods are also seen in other members of mollusc phylum - giant clams<sup>[[#Clams|[4]]]</sup>.
| |
| - | They are responsible for the stunning iridescent colours of the mantle, and may play a role in protecting against harmful UV and maximising capture of sunlight for the photosynthetic symbionts which live alongside the clam.
| |
| - |
| |
| - | ==Reflectins as novel polymers==
| |
| - |
| |
| - | Reflectins have unique properties in part due to their unique amino acid composition - residues which are normally common in proteins (alanine, isoleucine, leucine and lysine) are nowhere to be seen in any reflectins identified so far, whilst typically rare residues (arginine, methionine, tryptophan and tyrosine) make up ~57% of the protein.
| |
| - |
| |
| - | Kramer ''et al'' and Tao and DeMartini ''et al'' have demonstrated the remarkable capability of reflectin proteins to self assemble in vitro to create complex structures.
| |
| - |
| |
| - | ==Light and interference - The physics behind structural colour==
| |
| - |
| |
| - |
| |
| - | ====What is a Bragg reflector? - ''Thin film Interference''====
| |
| - | Bragg reflectors are structures of alternating layers of materials with different refractive indices. These structures dominantly reflect at a certain peak wavelength in relation to the individual separation of the layers. Each boundary layer exhibits partial reflection which through superposition lead to interference phenomena. The peak reflected wavelength is 4 times the spacing distance between layers whereby the path difference is such as to allow constructive interference. This is the fundamental principle behind thin film interference, responsible for the rainbow colours reflected by oil droplets on the surface of water and that present on soap films.
| |
| - |
| |
| - | <div id="diridescence"></div>
| |
| - |
| |
| - | ====What is dynamic iridescence and how does it work in squid? - ''Phosphorylation''====
| |
| - | Iridescence describes the material colour change as the viewing angle or the angle of incidence of light is varied. However dynamic iridescence, observed in certain squid genera is believed to be a result of neural control. Specifically, the application of the neurotransmitter Acetyl Choline (ACH) to fresh skin samples resulted in detectable post-translational modifications of the protein, namely [http://en.wikipedia.org/wiki/Phosphorylation phosphorylation]. It is believed that [http://en.wikipedia.org/wiki/Phosphorylation phosphorylation] of reflectin proteins cause changes in the chemical interactions within the nanoparticles reflectin forms in-vivo within the [http://en.wikipedia.org/wiki/Chromatophore#Iridophores_and_leucophores iridophore]. These changes subsequently induce an alteration in the volume of protein platelets of reflectin and critically the thicknesses of reflectin layers in the iridophore. The path difference between incident light on individual layers is thus altered resulting in a shift in peak reflected wavelength and therefore colour.
| |
| - |
| |
| - | ====What is known about reflectins?====
| |
| - |
| |
| - | No [http://en.wikipedia.org/wiki/Intron introns] were found in the reflectin genes when reflectin genes amplified from ''Euprymna scolopes'' genomic DNA were sequenced<sup>[[#Crookes|[2]]]</sup>.
| |
| - |
| |
| - | Reflectins are characterised by a highly unusual amino acid composition; tyrosine, methionine, arginine and tryptophan are relatively rare residues.
| |
| - |
| |
| - | ====What is known about the unusual amino acid content of reflectins?====
| |
| - |
| |
| - | ====What is known about the structure of the reflectin proteins?====
| |
| - |
| |
| - | ====What are the differences and conserved sequences between different reflectins and between reflectins in different species?====
| |
| - |
| |
| - | ====What in vitro experiments have been performed on reflectins?====
| |
| - | Reflectin that has been overexpressed in E. coli , purified and then refolded in vitro has been shown to be soluble in water and possesses self-assembly properties. Thin films of the protein have been made using a flow-coating technique, which have been shown to have the highest refractive index out of all other proteins measured to date. These films display Bragg interference phenomena, and have been shown to change colour across the visible spectrum when water or ethanol vapours are applied. It has been suggested that
| |
| - | this is due to expansion of the film under these conditions.
| |
| - | More intriguing self-assembly has been demonstrated by the spontaneous production reflectin diffraction gratings, with remarkably even spacing and very little sign of defects. These were made by simply dissolving reflectin in 1-butyl-3-methylimidazolium chloride (BMIM), casting the solution on a silicon wafer and immersing the resulting film in water.
| |
| - | Changing the rate at which the film was dipped in water varied the diffraction grating spacing.
| |
| - |
| |
| - | ====What are the properties of in vitro films of reflectin?====
| |
| - |
| |
| - | =='''References'''==
| |
| - | <div id="Kramer"></div>
| |
| - | [http://www.nature.com/nmat/journal/v6/n7/abs/nmat1930.html] Kramer ''et al.'' '''The self-organizing properties of squid reflectin protein''' Nature Materials 533-538 VOL6 JULY 2007
| |
| - | <div id="Crookes"></div>
| |
| - | [http://www.sciencemag.org/content/303/5655/235.short] Crookes ''et al.'' '''Reflectins: The Unusual Proteins of Squid Reflective Tissues''' SCIENCE 235-238 VOL303 9 JANUARY 2004
| |
| - | <div id="Morse"></div>
| |
| - | [http://www.sciencedirect.com/science/article/pii/S0142961209011442] Morse ''et al.'' '''The role of protein assembly in dynamically tunable bio-optical tissues''' Biomaterials 793-801 VOL31 FEBRUARY 2010
| |
| - | <div id="Clams"></div>
| |
| - | [http://www.publish.csiro.au/paper/ZO9920319.html]Iridophores in the mantle of giant clams
| |
| | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} |
 "
"