Team:Cambridge/Experiments/Low Level Expression
From 2011.igem.org
(→Low Level Expression) |
m (→Low Level Expression) |
||
| (41 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{Template:Team:Cambridge/ | + | {{Template:Team:Cambridge/CAM_2011_EXPERIMENT_HEAD}} |
=Low Level Expression= | =Low Level Expression= | ||
For our [[Team:Cambridge/Project/In_Vivo | in vivo]] work, we needed to be able to express reflectin at low levels, and control the level of expression reliably. Therefore, we expressed reflectin under an arabinose inducible promoter (pBAD) on a low copy plasmid ([http://partsregistry.org/Part:pSB3K3 pSB3K3]) in cells with a titratable response to arabinose. | For our [[Team:Cambridge/Project/In_Vivo | in vivo]] work, we needed to be able to express reflectin at low levels, and control the level of expression reliably. Therefore, we expressed reflectin under an arabinose inducible promoter (pBAD) on a low copy plasmid ([http://partsregistry.org/Part:pSB3K3 pSB3K3]) in cells with a titratable response to arabinose. | ||
| Line 8: | Line 8: | ||
*'''AraC''' protein included in the I0500 biobrick, which acts an a ''repressor''. | *'''AraC''' protein included in the I0500 biobrick, which acts an a ''repressor''. | ||
Therefore, the araC-pBAD system offers regulatable control of gene expression in the presence of the inducer and highly repressed in the absence of the inducer. | Therefore, the araC-pBAD system offers regulatable control of gene expression in the presence of the inducer and highly repressed in the absence of the inducer. | ||
| - | |||
| - | |||
| - | |||
| - | |||
AraC protein functions as a homodimer. The monomer possesses two domain - a dimerization domain that also binds arabinose and a DNA-binding domain. <br/>In the absence of arabinose, the repressor protein AraC in a dimerized form binds to the AraI1 operator site of pBAD and the upstream operator site AraO2. The resulting DNA loop sterically blocks access of RNA polymerase to the pBAD promoter, inhibiting transcription of the downstream genes. In the presence of arabinose, the monosaccharide binds to AraC and change its conformation such that it interacts with the AraI1 and AraI2 operator sites permitting transcription. | AraC protein functions as a homodimer. The monomer possesses two domain - a dimerization domain that also binds arabinose and a DNA-binding domain. <br/>In the absence of arabinose, the repressor protein AraC in a dimerized form binds to the AraI1 operator site of pBAD and the upstream operator site AraO2. The resulting DNA loop sterically blocks access of RNA polymerase to the pBAD promoter, inhibiting transcription of the downstream genes. In the presence of arabinose, the monosaccharide binds to AraC and change its conformation such that it interacts with the AraI1 and AraI2 operator sites permitting transcription. | ||
| - | + | Some of our data shed some light on the difficulties of characterizing the pBAD promoter. This information has been posted on the [http://partsregistry.org/Part:BBa_I0500:Experience Experience] page for I0500. | |
| - | + | ||
| - | *AraC represses transcription initiation at the pBAD promoter. | + | ==Arabinose System== |
| - | *AraC represses expression of its own. | + | The native arabinose system is used by E.coli to: |
| - | + | # take up L-arabinose from the growth medium by high-capacity and low-affinity trasporters: AraE and AraFGH; | |
| - | *AraC stimulates transcription of araE and araFGH genes | + | # convert intracellular arabinose into D-xylulose-5-phosphate in three reactions catalyzed by the products of the genes from the pBAD operon; |
| - | + | It is mainly controlled by the AraC repressor, which regulates expression of its own synthesis and the other genes of the arabinose system: | |
| - | *AraC represses expression of its own. | + | |
| + | ''In the absence of arabinose:'' | ||
| + | * AraC represses transcription initiation at the pBAD promoter. | ||
| + | * AraC represses expression of its own. | ||
| + | ''In the presence of arabinose:'' | ||
| + | * AraC activates transcription from the pBAD promoter. | ||
| + | * AraC stimulates transcription of araE and araFGH genes. | ||
| + | * AraC represses expression of its own. | ||
==E.coli Strain== | ==E.coli Strain== | ||
| Line 29: | Line 32: | ||
The linear induction with increasing arabinose concentration is not achievable in the standard E.coli strain, because the endogenous promoter which controls the araE gene is upregulated by an increasing concentration of arabinose. Thus, with higher level of the monosaccharide in the medium, more arabinose transporters are present in the plasma membrane and therefore the rate of uptake rises accordingly. | The linear induction with increasing arabinose concentration is not achievable in the standard E.coli strain, because the endogenous promoter which controls the araE gene is upregulated by an increasing concentration of arabinose. Thus, with higher level of the monosaccharide in the medium, more arabinose transporters are present in the plasma membrane and therefore the rate of uptake rises accordingly. | ||
| - | This induction of arabinose transporter can be circumvented by deleting the chromosomal araE gene and replacing it with a plasmid-borne copy of the araE under the control of a constitutive promoter. This is what has been done in the [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] strain of E.coli, which we used in our | + | This induction of arabinose transporter can be circumvented by deleting the chromosomal araE gene and replacing it with a plasmid-borne copy of the araE under the control of a constitutive promoter. This is what has been done in the [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] strain of E.coli, which we used in our experiment. |
==Constructs== | ==Constructs== | ||
| - | In this experiment we used confocal microscopy to compare distribution of Reflectin A1 when it is expressed at high and low levels in E.coli cells. In order to do so, we used four different plasmids with Reflectin A1 gene expressed under the control of the pBAD promoter, and their assembly is described in this [[Team:Cambridge/Experiments/Synthetic_Reflectin_PCR_and_Construction_of_GA1_to_6 | section]]. The [[Team:Cambridge/Experiments/Plasmid_Constructs | constructs]] we | + | In this experiment we used confocal microscopy to compare distribution of Reflectin A1 when it is expressed at high and low levels in E.coli cells. In order to do so, we used four different plasmids with Reflectin A1 gene expressed under the control of the pBAD promoter, and their assembly is described in this [[Team:Cambridge/Experiments/Synthetic_Reflectin_PCR_and_Construction_of_GA1_to_6 | section]]. The [[Team:Cambridge/Experiments/Plasmid_Constructs | constructs]] we worked on are the following: |
<center> | <center> | ||
{| | {| | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
|'''GA13''' | |'''GA13''' | ||
|Transcriptional fusion of Reflectin A1 and GFP on a high copy number plasmid [http://partsregistry.org/Part:pSB1A3 pSB1A3] | |Transcriptional fusion of Reflectin A1 and GFP on a high copy number plasmid [http://partsregistry.org/Part:pSB1A3 pSB1A3] | ||
| Line 51: | Line 48: | ||
==Observations== | ==Observations== | ||
| - | When reflectin was expressed on a low copy plasmid | + | When reflectin was expressed on a high copy plasmid (pSB1A3) all the cells contained inclusion bodies. On a low copy plasmid (pSB3K3) we saw fewer inclusion bodies. |
| + | |||
| + | <gallery widths=300px heights=300px> | ||
| + | Image:Cam_Reflectin-GFP-inclusionbodies.jpg | Reflectin-GFP on [http://partsregistry.org/Part:pSB1A3 pSB1A3](a high copy plasmid), induced with 1mM Arabinose. Fluorescence is localised in dots at the end of the cells, indicating that fusion protein is localised to insoluble inclusion bodies. | ||
| + | Image:Cam-Low-Copy-Ref-GFP.jpg | Reflectin-GFP on [http://partsregistry.org/Part:pSB3K3 pSB3K3] (low/med copy plasmid), induced with 1mM Arabinose. The fusion protein is distributed throughout the cytoplasm, indicating soluble protein is expressed. | ||
| + | </gallery> | ||
==Induction== | ==Induction== | ||
| - | Using a plate reader, we | + | Using a plate reader, we tracked the OD<sub>600</sub> and GFP fluorescence of ''E coli'' cells of strain BW27783, transformed with several constructs, after induction with arabinose. We took readings every 10 minutes for 20 hours. |
| - | + | The constructs used were: | |
| - | + | *[http://partsregistry.org/Part:BBa_J69511 BBa_J69511] for a baseline GFP signal. The promoter in this construct is not inducible with arabinose. | |
| + | *ReflectinA1 generator ([http://partsregistry.org/Part:BBa_K638101 BBa_K638101]) | ||
| + | *ReflectinA1-sfGFP generator ([http://partsregistry.org/Part:BBa_K638301 BBa_K638301]) | ||
| + | *TorA-ReflectinA1-sfGFP generator ([http://partsregistry.org/Part:BBa_K638403 BBa_K638403]) | ||
| + | all on the low-copy plasmid pSB3K3. | ||
| - | Regulation of the l-arabinose operon of Escherichia coli | + | We grew up liquid cultures overnight, diluted 10 times and then grew the cells up to an OD<sub>600</sub> of 0.5. We then transferred the liquid culture to a 96-well plate and induced with different concentrations of arabinose: 0M, 0.001mM, 0.01mM, 0.1mM, 1mM, 10mM. |
| - | + | ||
| + | With 4 constructs, 6 different arabinose concentrations, and 2 technical replicates, we used 48 wells in total. | ||
| + | |||
| + | Our initial results (see graphs below) show the following features: | ||
| + | *Induction appears to take about 1 hour. | ||
| + | *After 16-20 hours, all liquid cultures have OD<sub>600</sub> of 2.5 or more. This is consistent with our experience of growing overnight cultures under induction with arabinose: cells producing reflectin grow to a similar OD<sub>600</sub> to uninduced cells. | ||
| + | *The OD<sub>600</sub> readings of all of the uninduced cells (OM or 0.001mM arabinose) follow a very consistent pattern. The OD<sub>600</sub> rises steeply to about 3.0 after 800 minutes, and thereafter the OD<sub>600</sub> remains constant or slowly decreases. | ||
| + | *For the construct J69511 induction with arabinose had very little affect on OD<sub>600</sub> or GFP readings. | ||
| + | *For K638301, and K638403, cells grown in 0M or 0.001mM arabinose showed little fluorescence, those induced with 0.1mM or more showed strong fluorescence, and those induced with 0.01mM arabinose showed intermediate behaviour. | ||
| + | *For K638101 (ReflectinA1) growth was severely slowed after 1 hour. | ||
| + | *The cells producing K638301 (ReflectinA1-sfGFP) and K638403 (TorA-ReflectinA1-sfGFP), in contrast, show higher OD<sub>600</sub> readings when induced. | ||
| + | *The fact that growth is adversely affected by ReflectinA1 but not by the fusion ReflectinA1-sfGFP may be due to changes in solubility; sfGFP has been shown to improve the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2826880/ solubility] of proteins to which it is fused. | ||
| + | *These results cannot be fully explained without further work. For instance, the affect of reflectin on the OD<sub>600</sub> is not known and the increase in OD<sub>600</sub> of the cultures expressing ReflectinA1-sfGFP and TorA-ReflectinA1-sfGFP upon induction with arabinose cannot be explained. | ||
| + | |||
| + | |||
| + | |||
| + | OD<sub>600</sub> and GFP readings for construct J659411 after induction with arabinose: | ||
| + | <center> | ||
| + | <gallery widths='300px' heights='300px'> | ||
| + | File:Cam-1-BBa_J659411-OD.png | | ||
| + | File:Cam-1-BBa_J659411-GFP.png | | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | OD<sub>600</sub> and GFP readings for construct K638101 after induction with arabinose: | ||
| + | <center> | ||
| + | <gallery widths='300px' heights='300px'> | ||
| + | File:Cam-2-refA1-OD.png | | ||
| + | File:Cam-2-refA1-GFP.png | | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | OD<sub>600</sub> and GFP readings for construct K638301 after induction with arabinose: | ||
| + | <center> | ||
| + | <gallery widths='300px' heights='300px'> | ||
| + | File:Cam-7-refA1-gfp-OD.png | | ||
| + | File:Cam-7-refA1-gfp-GFP.png | | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
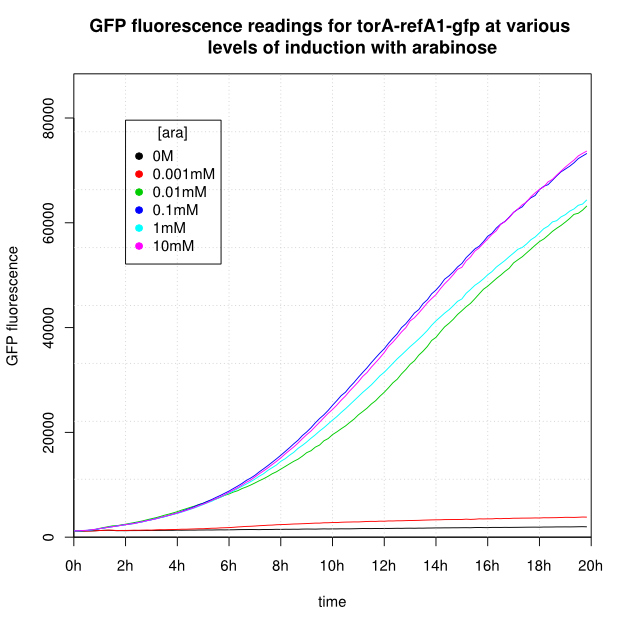
| + | OD<sub>600</sub> and GFP readings for construct K638403 after induction with arabinose: | ||
| + | <center> | ||
| + | <gallery widths='300px' heights='300px'> | ||
| + | File:Cam-8-torA-refA1-gfp-OD.png | | ||
| + | File:Cam-8-torA-refA1-gfp-GFP.png | | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | ==References== | ||
| + | #'''Schleif R. (2000)''' Regulation of the l-arabinose operon of Escherichia coli. ''Trends in Genetics'', 12:559-565. | ||
| - | |||
| - | {{Template:Team:Cambridge/ | + | {{Template:Team:Cambridge/CAM_2011_EXPERIMENT_FOOT}} |
Latest revision as of 00:39, 22 September 2011
Contents |
Low Level Expression
For our in vivo work, we needed to be able to express reflectin at low levels, and control the level of expression reliably. Therefore, we expressed reflectin under an arabinose inducible promoter (pBAD) on a low copy plasmid ([http://partsregistry.org/Part:pSB3K3 pSB3K3]) in cells with a titratable response to arabinose.
pBAD Promoter
The pBAD promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_I0500 I0500] is tightly controlled by two factors:
- L-arabinose monosaccharide taken up by the cell from the medium, which acts as an inducer.
- AraC protein included in the I0500 biobrick, which acts an a repressor.
Therefore, the araC-pBAD system offers regulatable control of gene expression in the presence of the inducer and highly repressed in the absence of the inducer.
AraC protein functions as a homodimer. The monomer possesses two domain - a dimerization domain that also binds arabinose and a DNA-binding domain.
In the absence of arabinose, the repressor protein AraC in a dimerized form binds to the AraI1 operator site of pBAD and the upstream operator site AraO2. The resulting DNA loop sterically blocks access of RNA polymerase to the pBAD promoter, inhibiting transcription of the downstream genes. In the presence of arabinose, the monosaccharide binds to AraC and change its conformation such that it interacts with the AraI1 and AraI2 operator sites permitting transcription.
Some of our data shed some light on the difficulties of characterizing the pBAD promoter. This information has been posted on the [http://partsregistry.org/Part:BBa_I0500:Experience Experience] page for I0500.
Arabinose System
The native arabinose system is used by E.coli to:
- take up L-arabinose from the growth medium by high-capacity and low-affinity trasporters: AraE and AraFGH;
- convert intracellular arabinose into D-xylulose-5-phosphate in three reactions catalyzed by the products of the genes from the pBAD operon;
It is mainly controlled by the AraC repressor, which regulates expression of its own synthesis and the other genes of the arabinose system:
In the absence of arabinose:
- AraC represses transcription initiation at the pBAD promoter.
- AraC represses expression of its own.
In the presence of arabinose:
- AraC activates transcription from the pBAD promoter.
- AraC stimulates transcription of araE and araFGH genes.
- AraC represses expression of its own.
E.coli Strain
In order to obtain a linear titratable relation between the concentration of arabinose in the medium and the level of reflectin expression, a special strain of bacteria [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] needs to be used.
The linear induction with increasing arabinose concentration is not achievable in the standard E.coli strain, because the endogenous promoter which controls the araE gene is upregulated by an increasing concentration of arabinose. Thus, with higher level of the monosaccharide in the medium, more arabinose transporters are present in the plasma membrane and therefore the rate of uptake rises accordingly.
This induction of arabinose transporter can be circumvented by deleting the chromosomal araE gene and replacing it with a plasmid-borne copy of the araE under the control of a constitutive promoter. This is what has been done in the [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783] strain of E.coli, which we used in our experiment.
Constructs
In this experiment we used confocal microscopy to compare distribution of Reflectin A1 when it is expressed at high and low levels in E.coli cells. In order to do so, we used four different plasmids with Reflectin A1 gene expressed under the control of the pBAD promoter, and their assembly is described in this section. The constructs we worked on are the following:
| GA13 | Transcriptional fusion of Reflectin A1 and GFP on a high copy number plasmid [http://partsregistry.org/Part:pSB1A3 pSB1A3] |
| GA14 | Transcriptional fusion of Reflectin A1 and GFP on a low copy number plasmid [http://partsregistry.org/Part:pSB3K3 pSB3K3] |
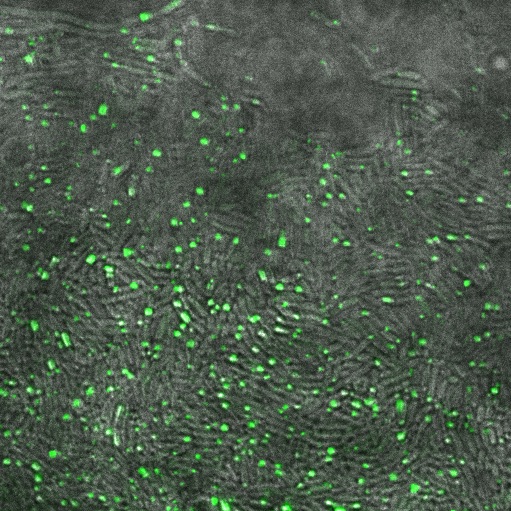
Observations
When reflectin was expressed on a high copy plasmid (pSB1A3) all the cells contained inclusion bodies. On a low copy plasmid (pSB3K3) we saw fewer inclusion bodies.
Induction
Using a plate reader, we tracked the OD600 and GFP fluorescence of E coli cells of strain BW27783, transformed with several constructs, after induction with arabinose. We took readings every 10 minutes for 20 hours.
The constructs used were:
- [http://partsregistry.org/Part:BBa_J69511 BBa_J69511] for a baseline GFP signal. The promoter in this construct is not inducible with arabinose.
- ReflectinA1 generator ([http://partsregistry.org/Part:BBa_K638101 BBa_K638101])
- ReflectinA1-sfGFP generator ([http://partsregistry.org/Part:BBa_K638301 BBa_K638301])
- TorA-ReflectinA1-sfGFP generator ([http://partsregistry.org/Part:BBa_K638403 BBa_K638403])
all on the low-copy plasmid pSB3K3.
We grew up liquid cultures overnight, diluted 10 times and then grew the cells up to an OD600 of 0.5. We then transferred the liquid culture to a 96-well plate and induced with different concentrations of arabinose: 0M, 0.001mM, 0.01mM, 0.1mM, 1mM, 10mM.
With 4 constructs, 6 different arabinose concentrations, and 2 technical replicates, we used 48 wells in total.
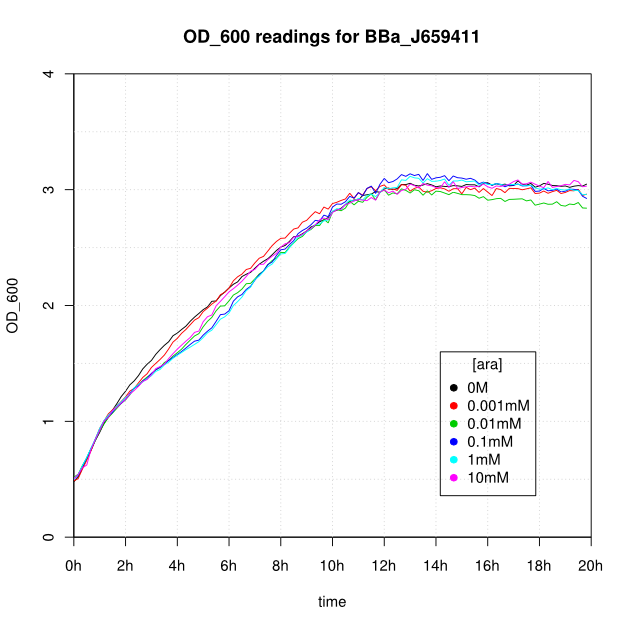
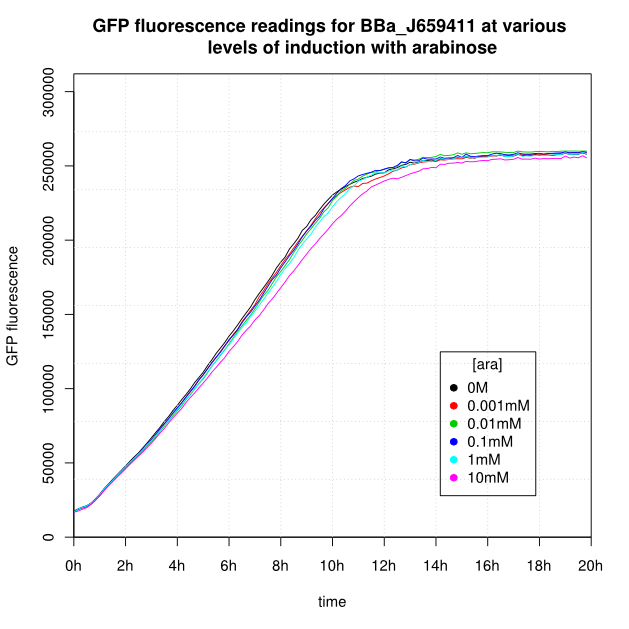
Our initial results (see graphs below) show the following features:
- Induction appears to take about 1 hour.
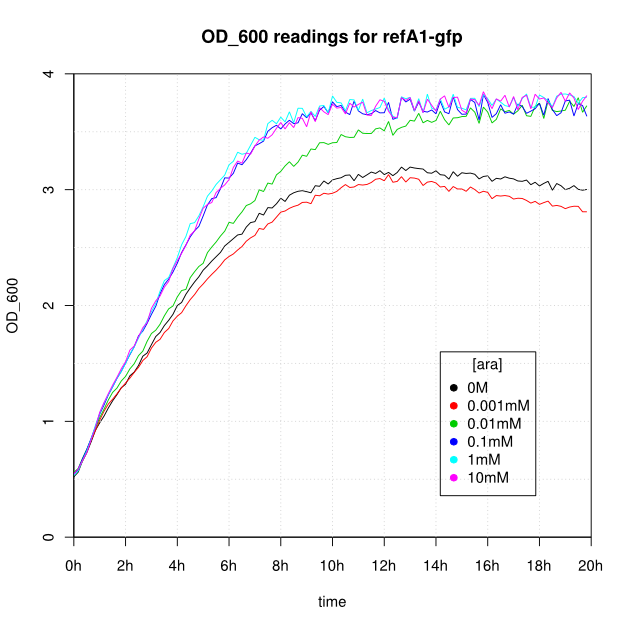
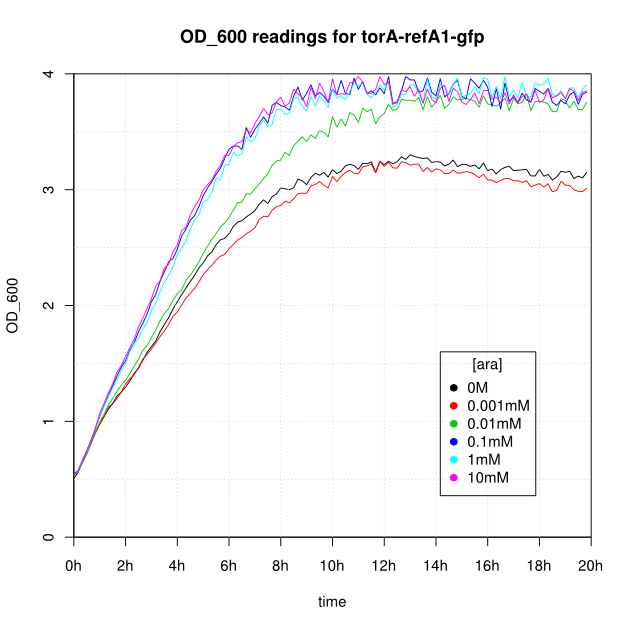
- After 16-20 hours, all liquid cultures have OD600 of 2.5 or more. This is consistent with our experience of growing overnight cultures under induction with arabinose: cells producing reflectin grow to a similar OD600 to uninduced cells.
- The OD600 readings of all of the uninduced cells (OM or 0.001mM arabinose) follow a very consistent pattern. The OD600 rises steeply to about 3.0 after 800 minutes, and thereafter the OD600 remains constant or slowly decreases.
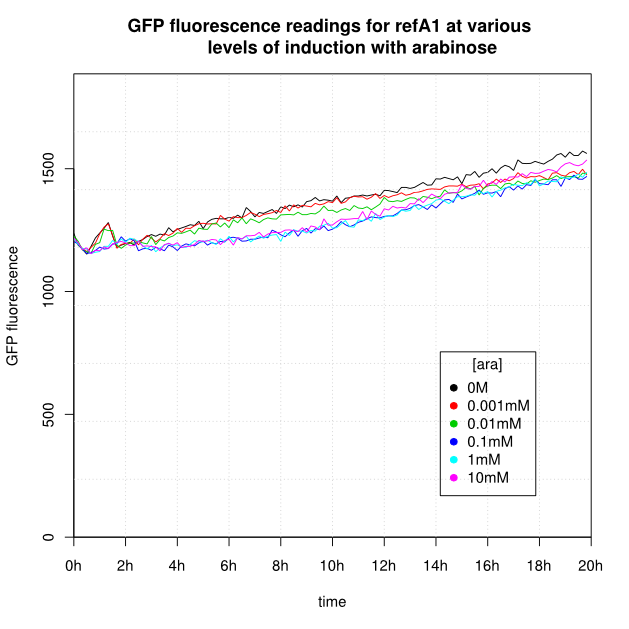
- For the construct J69511 induction with arabinose had very little affect on OD600 or GFP readings.
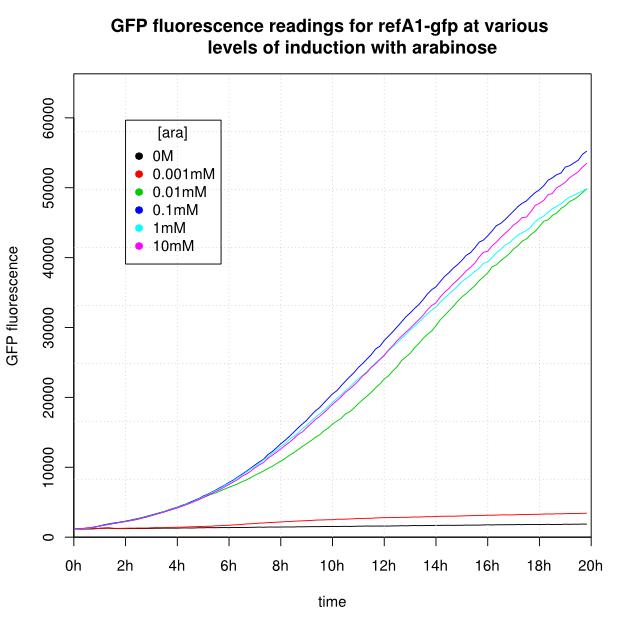
- For K638301, and K638403, cells grown in 0M or 0.001mM arabinose showed little fluorescence, those induced with 0.1mM or more showed strong fluorescence, and those induced with 0.01mM arabinose showed intermediate behaviour.
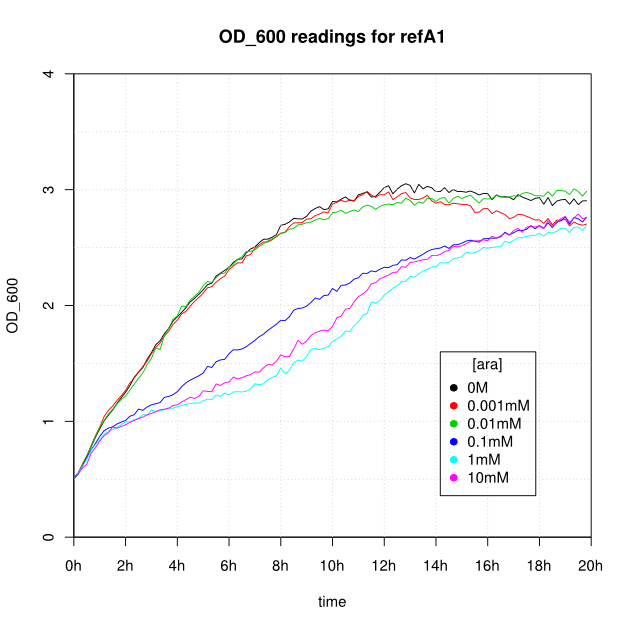
- For K638101 (ReflectinA1) growth was severely slowed after 1 hour.
- The cells producing K638301 (ReflectinA1-sfGFP) and K638403 (TorA-ReflectinA1-sfGFP), in contrast, show higher OD600 readings when induced.
- The fact that growth is adversely affected by ReflectinA1 but not by the fusion ReflectinA1-sfGFP may be due to changes in solubility; sfGFP has been shown to improve the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2826880/ solubility] of proteins to which it is fused.
- These results cannot be fully explained without further work. For instance, the affect of reflectin on the OD600 is not known and the increase in OD600 of the cultures expressing ReflectinA1-sfGFP and TorA-ReflectinA1-sfGFP upon induction with arabinose cannot be explained.
OD600 and GFP readings for construct J659411 after induction with arabinose:
OD600 and GFP readings for construct K638101 after induction with arabinose:
OD600 and GFP readings for construct K638301 after induction with arabinose:
OD600 and GFP readings for construct K638403 after induction with arabinose:
References
- Schleif R. (2000) Regulation of the l-arabinose operon of Escherichia coli. Trends in Genetics, 12:559-565.
Back to Experiments
 "
"