Team:Cambridge/Experiments/Initial Exercise Group control
From 2011.igem.org
(→PCR reaction) |
(→PCR reaction) |
||
| Line 33: | Line 33: | ||
* The graph presents accumulation of products with time in real-time PCR: | * The graph presents accumulation of products with time in real-time PCR: | ||
[[File:cam_PCR_graph_positive_control.jpg | centre | thumb | 400px | progress of PCR reaction]] | [[File:cam_PCR_graph_positive_control.jpg | centre | thumb | 400px | progress of PCR reaction]] | ||
| - | |||
| - | |||
====Gel Electrophoresis==== | ====Gel Electrophoresis==== | ||
Revision as of 14:36, 15 July 2011
Contents |
Positive Control Experiment
Construct Design
In the positive control experiment we replaced the Green Fluorescent Protein coding sequence in a pJS104 plasmid with a coding sequence for mRUBY, which is a Bright Monomeric Red Fluorescent Protein. The picture below shows a map of the modified plasmid.
Experiment
The experiment involved the same steps as preparation and expression of gene fusions of the three teams.
PCR reaction
- We amplified the mRUBY coding sequence and two arms of the plasmid in a PCR reactions. First, we performed a real-time PCR with Taq polymerase, but as most samples were poorly amplified, we decided to repeat the reaction with Phusion polymerase (protocol)
The three reactions performed ae the following:
- Reaction A
- 1μl primer ruby F (provided)
- 1μl primer ruby R (provided)
- 1μl mRuby as a template
- Reaction B
- 1μl primer Vector F (provided)
- 1μl primer B reverse (provided)
- 1μl pJS104 plasmid as a template
- Reaction C
- 1μl primer Vector R (provided)
- 1μl primer A forward (provided)
- 1μl pJS104 plasmid as a template
- The graph presents accumulation of products with time in real-time PCR:
Gel Electrophoresis
- Products of PCR reaction with Phusion polymerase were separated on 1% agarose gel according to the protocol. The location of bands corresponded to the rough estimate of predicted sizes of DNA fragments.
Gel Extraction of DNA
- We followed the protocol to extract and purify DNA.
Gibson Assembly
- We performed Gibson Assembly in order to obtain a complete plasmid with GFP gene replaced by mRUBY.
Transformation of E.coli Competent Cells
- We transformed competent E.coli cells with products of the Gibson Assembly, and after an overnight incubation at 37°C we examined colonies under the fluorescent microscope. We could see around 20 colonies on the plates, each emitting bright red light.
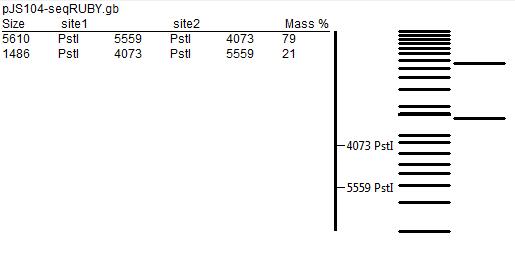
Digestion with Restriction Enzymes
In order to further test for the correct assembly, we performed restriction mapping of the plasmid. That involved extraction of DNA from E.coli colonies grown overnight in liquid medium using the QIAGEN MiniPrep Kit technique, followed by incubation with KpnI and PstI restriction enzymes. After that, we separated products of digestion by gel electrophoresis (using 1% agarose gel), but most probably due to poor quality of the gel and unfavourable concentration of the buffer used, the results did not match the predicted restriction map (presented below).
Transformation of Bacillus subtilis Competent Cells
As a further confirmation of correct assembly of the plasmid, we transfected competent Bacillus cells with purified DNA that had been isolated from E.coli.
 "
"