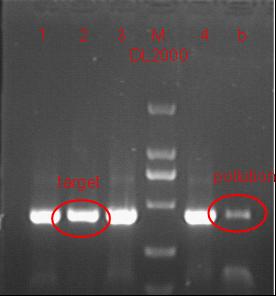
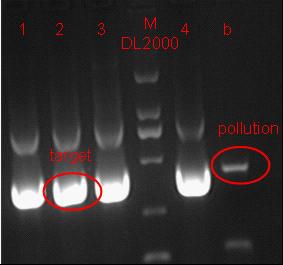
Team:SJTU-BioX-Shanghai/Notebook/Lablog2
From 2011.igem.org
|
|
Calendar
July
9/1/2011Zhangbo: 8.31 failed Nachte:Get Rosseta from WY,we contransformed 9/2/2011Zhangbo: 9.2 Buf 2ul DNA 16ul(plasmid) Nde1 1ul Xho1 1ul Total 20ul 37℃ 5hours 9/3/2011ZhangBo: 9.3 Binding the product together Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours 9/4/2011Chunying: Prepared the 4 flask containing 100ml medium to test the four kinds of bacterial(4S,4L,6S,6L luciferase that under the Amp promoter and the rare codon's tRNA is induced by IPTG. Zhangbo: Failed 9/5/2011test the 4AGG codon IPTG+Amp promoter system: 4C-co-transformed 4AGG codon IPTG+Amp-promoter system, IPTG+ 6C-co-transformed 6AGG codon IPTG+Amp-promoter system, IPTG+ 4S-only 4AGG codon IPTG pReporter, IPTG+ 6S-only 6AGG codon IPTG pReporter, IPTG+ The IPTG is added into the bacteria when its OD600 reached about 0.3. Take samples every one hour and last for 7 hours. ZhangBo: 9.5 RFP PCR PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WtRNA-1) 2/2ul Plasmid(WTM-9) 1ul KOD 1ul ddH2O 31ul Purified the product. DNA(above) 10ul plasmid 5ul PCR KOD buf 3ul dNTP 3ul Mg2+ 1.8ul KOD 1ul ddH2O 6.2ul Deal with the PCR result above with the DpnI 1ul enzyme(Fast Digest) into 50ul PCR result for 2 hours Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours 9/6/2011Test the effect of concentration of IPTG on the expression of luciferase. Try different concentration of IPTG: 0.1mM,0.2mM,0.3mM,0.4mM,0.5mM,0.6mM,0.7mM,0.8mM,0.9mM,1.0mM The bacteria used for test: 4AGG+(Amp-luciferase)+(IPTG-tRNA) 4AGG+(Amp-luciferase) without added tRNA The result indicated that the concentration of IPTG realy has effects on the expression of luciferase as well as the growth of bacteria. But we cannot find out there is no obvious relationship between the concentration and the amount of luciferase. But when the concentration is too high, the growth of bacteria is blocked. 9/7/2011ZhangBo: 9/8/2011ZhangBo: 9/9/2011Zhangbo: Co-transfer RFPW-pRS1.1 RFPM-pRS1.1 9/12/2011Try to introduce get wild type luciferase ( under the amp promoter) and the luciferase gene (amp promoter) that have 2 and 8 AGG codons after the initial codon. Still use the point mutation to conduct it. We try different PCR conditions . 1)20 cycles, 60℃ for annealing, 1ul template+2ul primers ( in 50ul system). 2)20 cycles, 60℃ for annealing, 1ul template+3ul primers ( in 50ul system). 3)30 cycles, 55℃ for annealing, 1ul template+1ul primers ( in 50ul system). Zhangbo: 9.12 Co-transfer Again Choose the right colonies and grow it in the correct LB liquid medium 9/13/2011We get the wild type luciferase gene (under Amp promoter) by mutation. But the genes that have 2 or 8 AGG codons are not gotten this time. Redo the point mutation PCR to get the luc genes that have 2 or 8 AGG codons. Zhangbo: 9.13 Choose the right colonies and grow it in the correct LB liquid medium of the Co-transfer Again Grow the first time for more time. 9/14/2011We failed again to have the luciferase that have 2 or 8 AGG and trancripted after Amp promoter. At the same time, we get the primers to get the luciferase gene that have 2 or 8 AGG codons but under the T7 promoter. Zhangbo: 9.14 RFPW turn red RFPM no result. 9/16/2011We find out that the tRNAAsp-TAG and its corresponding aaRS worked as we excepted. The reporter we use is RFP(inserted 6 AGG). Now we want to test if the aaRS can still charge the tRNA when the anticodon of the tRNA is mutated to UAG. So we design the primers to insert a amber codon after the luciferase gene's initial codon. 9/18/2011We use point mutation to insert one stop codon after the initial codon of luciferase gene. And later in the day we transfer the plasmid into DH5α. 9/19/2011The bacteria that has the mutated luciferase(having one stop codon after its initial codon)has grown on the plate.We Pick monoclonal colony and extract the plasmid. Then we sent the plasmid to be sequenced.
9/20/2011The sequencing result showed that we have successfully insert one stop codon after luciferase gene's initial codon. Then we transformed the plasmid into Er2566 along and co-transformed it with the aaRS+tRNA pairs that we constructed(the tRNA's anticodon is mutated to correspond to the stop codon) 9/21/2011We get the colonies:1.only have the pReporter(one stop codon after luciferase's initial codon) 2.have the pReporter and the pSwitch(the aaRS and tRNA we constructed) We incubated the monoclones into the medium. The sequencing of 4AGG(T7)and 6AGG(T7) indicated that the 4AGG(T7) plasmid is right but the 6AGG(T7) plasmid is wrong. Today, we began to reconstruct the 6AGG(T7) plasmid. 9/22/2011Test if the pReporter we constructed can turn up the expression of luciferase that has one stop codon after the initial codon. The method we used to test the luciferase's activities is the same with we used to use. The result indicated that the pSwitch really worked !!!! And the expression of luciferase is greatly increased when there are the aaRS+tRNA. The new 6AGG(T7) plasmid is transformed to DH5α this morning and we pick the monoclonies late in the day. 9/23/2011Extract 6AGG(T7) plasmid and send it to be sequenced. 9/24/2011The result of 6AGG(T7)'s sequencing indicated that the luciferase is inserted 4 AGG codons after the gene's initial codon instead of 6 AGG codons. By examining the previous work, we found out that the primers used for getting the 6AGG(T7)plasmid is wrong!!! It is the same with the primers used for 4AGG(T7). We syntheties the new primers as soon as possible. 9/26/2011We constructed the 6AGG(T7) plasmid again. 9/27/2011Reporter for Quantitative Analysis + tRNA Modulator. 2AGG(T7 promoter)+tRNA(lac promoter). 8AGG(T7 promoter)+tRNA(lac promoter). Every test point have three parallele goups 9/28/2011We finaly get the right 6AGG(T7) plasmid. And we transform it solely and co-tranRMAsform it with the pSwitch(IPTG+tRNA). 9/29/2011Reporter for Quantitative Analysis + tRNA Modulator. 4AGG(Amp promoter)+tRNA(lac promoter). 8AGG(Amp promoter)+tRNA(lac promoter). Every test point have three parallele goups. 9/30/2011Reporter for Quantitative Analysis + tRNA Modulator. 4AGG(T7 promoter)+tRNA(lac promoter). 6AGG(T7 promoter)+tRNA(lac promoter). Every test point have three parallele goups Reporter for Quantitative Analysis + aaRS Modulator one stop codon after the initial codon in luciferase+aaRS modulator Every kind of bacteria has three parallel groups |
 "
"