Team:SJTU-BioX-Shanghai/Notebook/Lablog1
From 2011.igem.org
|
|
Calendar
July
8/1/2011Phoenie: re-test the strains. I find that the PCR with bacteria culture and plasmid gives different result. PCR with bacteria culture shows that the AGG4 primers can also give positive result and PCR with plasmid doesn't Pony: Extracted pTrc99B from 4 tubes. Only one is succeed. Then, Bear amplified lacI gene+Trc promoter from pTrc99B and inserted the DNA fregment into plasmid pACYC184 by overlapping PCR. 8/2/2011Phoenie: The seller called me and told me that he had vacation in July and today is his first working day. The enzyme will arrive TOMORROW. next time we must order the enzyme when there is at least 10 mul left. 太坑爹了! 8/3/2011Chunying:Use the optimized primers to amplify the wild luc gene,failed. Try different proportion of the template and the primer(template 2ul,primer 10p,20p,30p) to amplify the gene, the result will come out tomorrow. Zhangbo: We have changed the reporter gene into RFP and design the new primer for the PCR. I have a attempt towards the tRNA mutation Nachte: Our problem is that a right fragment with right extending primers just can not be insert into that danmed plasmid. 8/4/2011Chunying:Different proportion of the template and primers did not result in successful amplification of the wild luc gene. The third amplification got the luc gene. Insert the wild luc gene into the pReporter(replace the amp gene). Transform the new pReporter plasmid into the cell and spread bacteria on the plate.(total 4 group, PCR at four different annealing temperature-50,53,57,60) 8/5/2011Zhangbo: I have done the PCR of the tRNA mutaion and transfer the plasmid into DH5a. And utilizing the restriction enzyme to cleave the pET28a-amp and the trpRS product. Chunying:The four transformations failed.Amply the wild luc gene again. 8/6/2011Zhangbo construction of the tRNA with the promoter and the terminator, I use the PCR to get this parts. PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WtRNA-1) 2/2ul Plasmid(pBRIVTC3.4) 1ul KOD 1ul ddH2O 31ul Program: 94 5min 94 30s 55 30s 68 30s 31cycle 68 7min Purify the DNA by TianGen Solution. Overlapping PCR DNA(above) 10ul pACYC184 5ul PCR KOD buf 3ul dNTP 3ul Mg2+ 1.8ul KOD 1ul ddH2O 6.2ul Program: 94 5min 94 30s 55 30s 68 4min30s 31 cycle 68 7min Instruction of the trpRS by Nco1 and Nde1 PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(trpRS) 2/2ul Plasmid(pRS1.1) 1ul KOD 1ul ddH2O 31ul Purified the PCR product by Tiangen PCR purification Kit Restriction enzyme Buf 2ul DNA 16ul(plasmid) Nde1 1ul Nco1 1ul Total 20ul Deal with the PCR result above with the DpnI 1ul enzyme(Fast Digest) into 50ul PCR result for 2 hours Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Bind the pET28a and PCR product PEG4000 1.5ul Buf 1.5ul Plasmid 3ul Pcr product 6ul Lingase 1ul ddH2O 2ul Total 15ul Program 22℃ 2 hours 72℃ 10min Transfer the product into the Dh5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Bind the pET28a and PCR product 8/7/2011Phoenie: Last night the plate which we spreat bacteria on have colonies on it but this morning the postive test showed that none of them contains the pET28a-luc plasmid. Parrot suggests that we can change the amount of plasmid and PCR product in each insertion reaction. And we re-cloned the luciferase with both sets of homogenous arms and performed a series of reaction with gradient plasmid and PCR product concertration and purified, transformed the insertion product. We also cultured the strain which was co-transformed with the pSwitch-arg and pReporter-AGG6 last night and the positive strains with pReporter-TAG BTW, the internet is so poor in the office today, I can hardly open any website except the homepage of Baidu. __....__
.-~~/ \__/ \~~-. If only the web can be a little faster~
/_/``\__/ \__/``\_\.--. / /
/ \__/ \__/ \__/ \ o`. /
`==/\__/__\__/__\__/__\__/\`'--' _______ /
~/__/__/^^^^^^^^\__\__\~ |_______|
8.7 Zhangbo: PCR testing on the Plasmid(WtRNA)(purified by tiangen Kit) PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul PCR testing on the Plasmid(trpRS-lingase)(purified by tiangen Kit) PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul 8/8/2011Phoenie: the socond co-transform is success and begin to insert by two step method 8.8 Zhangbo I choose the WTM-2 and the WRS-1,2 for the sequencing by JieLi corp. Nachte: Another cycle of cloning,we transformed ArgW-pTrc99b. 8/9/20118/10/2011Chunying:Divide the wild luciferase into two parts then amplify and insert them respectively. Today, inserted the latter part of the luc gene into the plasmid(pET28a-Amp) and transformed the new plasmid into Dh5a. In case that the luciferase will not be the best choice for our reporter gene, we use beta-lactamase as the other candidate for the reporter gene( because of its easier conduction and cheaper detection). Today we use the primer(R&L 004/005) to introduce mutation into the beta-lactamase gene (in its signal peptide)on the pET28a-Amp and transformed the new plasmid into Dh5a and spread bacterial on the Kana plate. Zhangbo: In this morning, the result of the sequencing of the samples has arrived by E-mail. The result of the tRNA lose a base in a very important part which may lead to the failure of the expression of the tRNA I want. So i check my primer and try for another time in this week. Nachte: Vaccinated several clonies of ArgW-pTrc99b. 8/11/2011Chunying: The bacterial that has half of the wild luciferase gene has grown on the plate. Negative screening for the stains. ZHangbo: 8.11 Instruction of the tRNA with the promoter and the terminator, I use the PCR to get this parts. PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WtRNA-1) 2/2ul Plasmid(pBRIVTC3.4) 1ul KOD 1ul ddH2O 31ul Program: 94 5min 94 30s 55 30s 68 30s 31cycle 68 7min Purify the DNA by TianGen Solution. Overlapping PCR DNA(above) 10ul pACYC184 5ul PCR KOD buf 3ul dNTP 3ul Mg2+ 1.8ul KOD 1ul ddH2O 6.2ul Program: 94 5min 94 30s 56 30s 68 4min30s 31 cycle 68 7min Nachte:Extracted plasmid ,verificating by PCR,and sending for sequencing. Alfred:Design the primers needed: For T7 promoter plus terminate containing MCS T7-F:5'-GCGGTGCGGACTGTTGTATTAATACGACTCACTATAGGGG-3' T7-R:5'-GATCGTGCTCCTGTCGTTGCCGGAATTCCAAAAAACCCCTCAAGAC-3' For truncated AspRS (without N-terminal anti-codon binding domain) TDRS-F: 5’-CATGCCATGGATGTTCTGCCGCTTGACTCT-3’ For AspV gene(tRNAAsp) AspV-F: 5’-CCACCATACCCACGCCGAAACCTAAAAATAGCGACTTGGGC-3’ AspV-R: 5’-TTGCCGTTACGCACCACCCTTGAACAGAGAAATGGTGGAA-3’ For mutation of AspV gene (GTC→CCT) AspV-M-F: 5’- AGTCGGTTAGAATACCTGCCTCCTACGCAGGGGGTCGC -3’ AspV-M-R: 5’- GCGACCCCCTGCGTAGGAGGCAGGTATTCTAACCGACT -3’
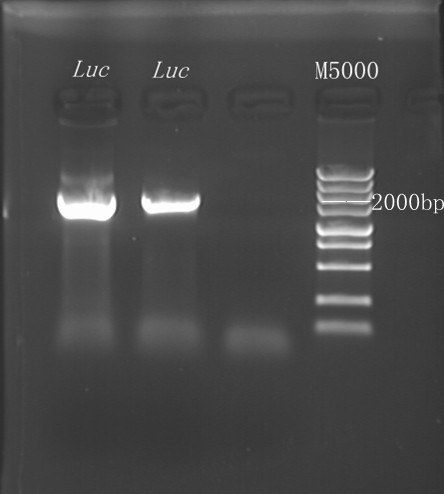
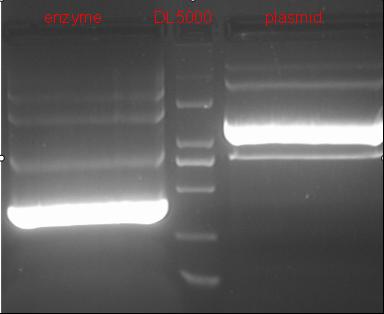
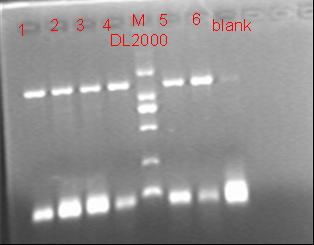
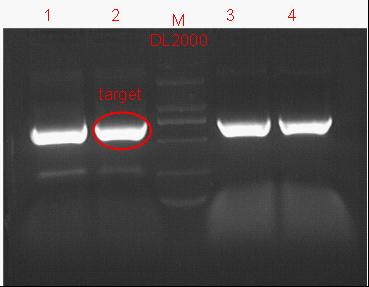
8/12/2011Phoenie: The colony PCR shows that we may get the plasmid with half of the luciferase gene on it. we culture the colony in liquid medium. However other method turned out unsuccessful. Zhangbo:8.12 The result of the WRS by T7 promoter is not correct compared with the trpRS in my database. Instruction of the trpRS by Nco1 and Nde1 PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(trpRS) 2/2ul Plasmid(pRS1.1) 1ul KOD 1ul ddH2O 31ul Purified the PCR product by Tiangen PCR purification Kit Restriction enzyme Buf 2ul DNA 16ul(plasmid) Nde1 1ul Nco1 1ul Total 20ul Deal with the PCR result above with the DpnI 1ul enzyme(Fast Digest) into 50ul PCR result for 2 hours Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Nachte:I went to the Kendo training,so it's midnight when I heard that Pony didn't gain GFP and RFP fragments with my primers.According to 8.13 is my birthday , burning midnight oil in the lab should be a awfully good idea of celebrating. 8/13/2011Phoenie: We extract the pET28a-halfluc and insert the front part of wt luc and 4AGG luc into the plasmid. Zhangbo: 8.13 PCR testing on the Plasmid(WtRNA)(purified by tiangen Kit) PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul I choose both for sequencing. Bind the pET28a and PCR product PEG4000 1.5ul Buf 1.5ul Plasmid 3ul Pcr product 6ul Lingase 1ul ddH2O 2ul Total 15ul Program 22℃ 2 hours 72℃ 10min Transfer the product into the Dh5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Bind the pET28a and PCR product Nachte:My spectulation is right~ Only a little change of the PCR program brought our target to me= 3= Well,frankly,my dear,it's my fault to create such a pair of abnormal temperature requiring primers. Alfred:Amplify T7 promoter plus terminate containing MCS from pET-28a plasmid. Reaction mixture: 10× PCR buffer for taq 5μl 2.5mM dNTP Mixture 5μl 25mM MgCl2 3μl TaKaRa Taq 0.5μl T7-F+T7-R 2+2μl pET-28a plasmid 2μl ddH2O up to 50μl After extract the PCR product, I introduced the T7 promoter plus terminate containing MCS into pACYC-184 plasmid (overlapping PCR method). Reaction mixture: 10× PCR buffer for KOD 3μl 2mM dNTP Mixture 3μl 25mM MgSO4 1.8μl KOD - Plus 0.3μl T7 segment 5μl pACYC-184 plasmid 5μl ddH2O up to 30μl After overlapping PCR, digest the PCR product with DpnI enzyme for eliminate the template plasmid. Subsequently, transform it into DH5α competent cell. 8/14/2011Phoenie: the result of positive test PCR is quite pulsing. It seems that the front part of the luciferase gene has not been inserted into the plasmid pET28a-halfluc right. So several other colonies are picked and cultured for further test in hope of getting the right strains Nachte:God-Wang send me a pair of ultra homologous and he guaranteed that they work well.I hope so@(ω)@ Do a new cycle of construction. Alfred:Choose eight colonies from yesterday’s plant and incubate them in LB liquid medium. 3 hours later, conduct a PCR test for validating whether the T7 promoter plus terminate containing MCS is introduced into pACYC-184 plasmid. Reaction mixture: 10× PCR buffer for taq 2μl 2.5mM dNTP Mixture 2μl 25mM MgCl 1.2μl TaKaRa Taq 0.3μl T7-F+T7-R 1+1μl colony 1μl ddH2O up to 30μl I found that all the colony is correct, so I choose one colony to incubate amplifiedly and extract plasmid, named as pACYC-T7. Amplify AspV gene from E.coli genome. Reaction mixture: 10× PCR buffer for taq 5μl 2.5mM dNTP Mixture 5μl 25mM MgCl2 3μl TaKaRa Taq 0.5μl AspV-F+ AspV-R 2+2μl E.coli genome 2μl ddH2O up to 50μl Introduce AspV gene into pACYC-T7 through overlapping PCR. After overlapping PCR, digest the PCR product with DpnI enzyme for eliminate the template plasmid. Subsequently, transform it into DH5α competent cell. 8/15/2011Phoenie: Send pET28a-halfluc and several other stains to the company for sequencing. extract the plasmid form the strain that may contain pReporter-wtluc pReporter-4AGG wtluc. The following figure shows the PCR test result. Zhangbo: 8.14 / 8.15 PCR testing on the Plasmid(trpRS-lingase)(purified by tiangen Kit) PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul I choose the WRS-3,4 for sequencing. Nachte:YOOOOOO!The god's creator doesn't work as well! Alfred:There were several colonies on the yesterday’s palnt. I incubated them in LB liquid medium and extract plasmid named as pACYC-T7-AspV. Then I did the PCR to validate its correctness. Amplify the truncated AspRS for E.coli genome with the primer TDRS-F+ TDRS-R. 8/16/2011Phoenie: Today we test the luciferase activity of E.coli strain ER2566 which contains pSwitch and pReporter to test the argW system and suppressor system. When the OD600 of the bacteria culture reaches 0.8, we expose the experiment groups under UV light in the sterilized petri dish for 20s.(the power of the UV lamp is 20W; distance between the lamp and petri dish is around 35cm). Then the exposed culture liquid is collected and cultured for another 2 hours. Then culture is centrifuged by 8000 r/min for 3min and resuspended with 2ml 0.85% sterilized pre-cooled NaCl solution. The OD600 of each suspension is measured The bacteria is ultrasonic disintegrated for 2 min, with 3s pulse for every 3s work. The sonicated bacterial cells is centrifuged by 10000 r/min for 1min to get rid of the cell debris. Then we measure the luciferase activity in each sample (mix 100 mul supernatant with 100 mul luciferin substrates). The result is somehow confusing group luciferase activity/OD066 AGG6 experiment group ~11,000,000/3 AGG6 control group ~11,000,000/3 TAG2 experiment group ~2,000/3 TAG2 control group ~1,000/3 Phoenie: the sequencing result shows that the AGG4 mutation has been constructed correctly. ZHangbo: 8.16 In the result of the WTM-1234, i find that the base is still missing and i check the sequence of the tRNA in my database which lead to a sad truth that the the missing is caused by myself so the construction of the tRNA successes. Nachte:Start to build pReporter~No one has experience with linker design,so we have to begin with a terrible high annealing temperature-72℃. The PCR result is horrorable,there are so many mispriming bands that Gel Extraction should be the best choice. 8/17/2011Phoenie: Today we test the luciferase activity of the DH5a stains which contains only pReporter-6AGG and pReporter-4AGG respectively. We culture the bacteria until the OD600 is around 0.8 and directly measure its luciferase activity, without UV exposure and additional 2-hour culture. The result shows that the relative enzyme activity of stains which contains only pReporter-6AGG or pReporter-4AGG is lower than those that contains both pSwitch and pReporter. The strain containing 4AGG has higher activity than that containing 6AGG. ZHangbo: 8.17 From the work result and the discussion of 8.17, the program of mine changed and find out several mistakes in my program into the instruction of the T7 promoter from the pET28a to the pACYC184 plasmid by the overlapping PCR. Redesign the primer WRS-2 for the mistake on the Nco1 restriction site which has a ATG within. Design the T7-1 primer. Nachte:Holly shit, the part of fusion protein can't be inserted into the vector too,regarding which our plan of overlapping PCR may be not a good way. Alfred:The mutation of pACYC-T7-AspV was failed. Try again with yesterday’s protocol. In the contrast, the linkage of TDRS segment and pET-28a was successful. Do the colony PCRs using T7-F and T7-R. Finally, extract the plasmid named pET-TDRS. 8/18/2011Phoenie: We test the co-transfered colonies by colony PCR. We use the strains without pSwitch-ArgW as negative control. All the five colony we pick give positive result. (The pSwitch-ArgW negative control shows a band that is much weaker than the tested samples.) Zhangbo: 8.18 Overlapping PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(T7-1) 2/2ul Plasmid(pET28a) 1ul KOD 1ul ddH2O 31ul Program: 94 5min 94 30s 55 30s 68 30s 31cycle 68 7min DNA(above) 10ul Plasmid(WTM-9) 5ul PCR KOD buf 3ul dNTP 3ul Mg2+ 1.8ul KOD 1ul ddH2O 6.2ul Program: 94 5min 94 30s 55 30s 68 4min30s 31 cycle 68 7min Nachte:In this week's gruop meeting ,we made a difficut decision-change to digestion,every step!I spend the whole afternoon to create the new primers,as the sites of EcoRI and XholI would be added to the 5'end.At the midnight,thanX to my bored feeling,I made the last attempt to get pTEnd by overlapping PCR,with a new program: 65 30s 68 5min*10cycles 60 30s 68 5min*10cycles 55 30s 68 5min*10cycles Finally,I got it~ Alfred:There were so many colonies of pACYC-T7-AspV mutation on the plant, so that I suspected the DpnI did not work. So I conducted 16 colony PCRs, and finally extracted eight tubes of plasmid to sequence. The align result was followed. So I confirmed the mutation of pACYC-T7-AspV was done, which named pACYC-T7-AspV(CCT).
8/19/2011We test different strains under the following conditions: co-transformed 4 AGG codon exposed by UV; co-transformed 4 AGG codon non-UV; only 4 AGG codon pReporter exposed by UV; only 4 AGG codon pReporter non-UV; co-transformed 6 AGG codon exposed by UV; co-transformed 6 AGG codon non-UV; only 6 AGG codon pReporter exposed by UV; only 6 AGG codon pReporter non-UV; We take samples after UV exposure every half an hour and measure the OD600 of the 5ml bacteria culture, OD600 of the 2ml suspension NaCl solution, DNA concentration of lysis solution, RLU after mix 100 mul solution and 100mul luciferin substrate. Zhangbo: 8.19 Transfer the product into the Dh5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Bind the pET28a and PCR product Nachte:We are always too simple to be a qualified reseacher! I t's easy to know what is digestion from the text book but setting a digestion primer is not a piece of cake.Nearly every new primer should be rewrited%> <% Alfred:Design primers: RFP - 1F: 5’- GGAATTCCATATGGCTTCCTCCGAAGACG-3’ RFP - 1R: 5’- CCGCTCGAGTTAAGCACCGGTGGAGTGAC-3’ RFP-2F: 5’- GGAATTCCATATGAGGAGGAGGAGGAGGAGGGCTTCCTCCGAAGACG-3’ Amplify the RFP gene and RFP mutation(AGG) from the plasmid on the 2011 Kit Plant (use RFP-1F+ RFP-1R and RFP-2F+ RFP-1R). Then do the PCR product purification. Digest the RFP segment and RFP mutation(AGG) with NdeI and XhoI at the 37℃ for 5 hours. So did the pACYC-T7-AspV(CCT), then linked the RFP segment and RFP mutation(AGG) and pET-28a with T4 ligase. Subsequently, transform it into DH5α competent cell.
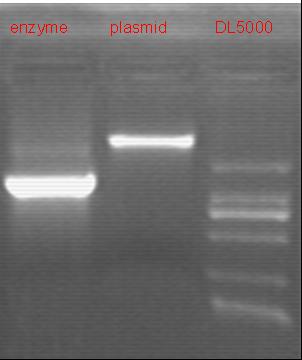
8/20/2011We discuss the result of the experiment and prepare the medium and other equipments. Zhangbo: 8.20 8.19 failed. The result of the WRS by T7 promoter is not correct compared with the trpRS in my database. Instruction of the trpRS by Nco1 and Nde1 PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WRS-2) 2/2ul Plasmid(pRS1.1) 1ul KOD 1ul ddH2O 31ul Purified the PCR product by Tiangen PCR purification Kit Restriction enzyme Buf 2ul DNA 16ul(plasmid) Nde1 1ul Nco1 1ul Total 20ul Nachte:The sequencing result of pTEnd produced(using primers of our own),not untill then I recognized that I had left the RBS before the ArgW sequence.It's hard to say whether that bug will influence the tRNA-Arg(AGG) expression...BTW,all depend on the contransformation result of pTEnd and team-1's reporter. Alfred:Do the colony PCRs for what I transformed yesterday (RFP:6 colonies, RFP-mutation:6 colonies). Subsequently, I extracted the plasmids for the positive result. These two plasmid were titled pACYC-RFP-AspV(CCT) and pACYC-RFP(AGG)-AspV(CCT). All the plamids needed, including pET-TDRS, pACYC-RFP-AspV(CCT) and pACYC-RFP(AGG)-AspV(CCT), were sequenced by invotrogen.
8/21/2011We optimize the experiment protocal and test the background RLU by measure the strain with only 4AGG pReporter and co-transformed 4AGG UV system. Zhangbo 8.21 binding the product together Bind the pET28a and PCR product PEG4000 1.5ul Buf 1.5ul Plasmid 3ul Pcr product 6ul Lingase 1ul ddH2O 2ul Total 15ul Program 22℃ 2 hours 72℃ 10min Transfer the product into the Dh5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Bind the pET28a and PCR product Alfred:Transform pACYC-RFP-AspV(CCT) into ER2566 competent cell. Transform pACYC-RFP(AGG)-AspV(CCT) into ER2566 competent cell. Co-transform pACYC-RFP(AGG)-AspV(CCT) and pET-TDRS into ER2566 competent cell. 8/22/2011Zhangbo: 8.22 8.21Failed. WtRNA introduction to pACYC184-T7 Overlapping PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WtRNA) 2/2ul Plasmid(WTM-9) 1ul KOD 1ul ddH2O 31ul Program: 94 5min 94 30s 55 30s 68 30s 31cycle 68 7min DNA(above) 10ul Plasmid(pACYC184-T7) 5ul PCR KOD buf 3ul dNTP 3ul Mg2+ 1.8ul KOD 1ul ddH2O 6.2ul Program: 94 5min 94 30s 55 30s 68 4min30s 31 cycle 68 7min Deal with the PCR result above with the DpnI 1ul enzyme(Fast Digest) into 50ul PCR result for 2 hours Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Nachte:New program!Referencing to the team-1's findings,besides changing of pReporter(AGG)'s promoter,we made another linker include TAG,which must be very strict as an obstacle. Alfred:Choose three colonies for each to incubate in 5ml LB liquid medium and induce expression with IPTG (1mM). 24 hours later, centrifugate the bacterial suspension and get the deposition. Observe under nature light and UV.
8/23/2011Zhangbo: 8.23 PCR testing on the Plasmid(WtRNA)(purified by tiangen Kit) PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul Sequencing Nachte:Ordering primers for TAG and AGG-digestion. 8/24/2011We test the the lac system (with 4 AGG codon and 6 AGG codon respectively)for the fist time by compare the differences between bacteria with only pReporter and both pSwitch and pReporter under stimulated condition. The result is that the OD600 of the culture stop rising very early when it is around o.6 for bacteria with pReporter and around 0.9 for co-tranformed bacteria 8/25/2011We test the UV system by testing: the co-transformed 4AGG UV system UV+ only 4AGG pReporter UV+ the co-transformed 4AGG UV system UV- Taking sample: immediately before UV exposure,0.5h 1h 1h20min 1h45min 2h15min 3h 4h 5h after UV exposure Nachte:The new primers for TAG have arrived,so we did a new cycle of linking GFP with RFP.From the process of gel extraction we found that linker-TAG works much better under a fixed temperature between the two Tm of primer-GFP-F and primer-RFP-R in which there is a big gap. 8/26/2011Zhangbo: 8.26 I find out that the DH5a has been polluted by a plasmid which caused the poor result of the link. Try another Dh5a and try again. Instruction of the trpRS by Nco1 and Nde1 PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(WRS-2) 2/2ul Plasmid(pRS1.1) 1ul KOD 1ul ddH2O 31ul Purified the PCR product by Tiangen PCR purification Kit Restriction enzyme Buf 2ul DNA 16ul(plasmid) Nde1 1ul Nco1 1ul Total 20ul Nachte:A big wave of primers arrived!As it's hard to insert such a long fragment into the vector,both overlapping extension and digestion would be taking in acount,and there are the primer couples: GFPoverR(3*AGG)+GFPoverF GFPdigR(TAG)+GFPoverF GFPoverR(3*AGG)+GFPdigF GFPdigR(TAG)+GFPdigF RFP(3*AGG)+RFPdigR RFP(TAG)+RFPdigR
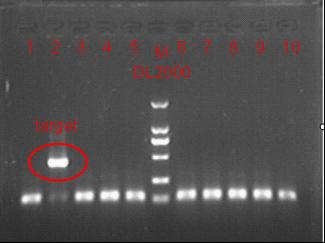
GFP part 3ul RFP part 3ul primers 3/3ul dNTPs 5ul 10*buffer 5ul Mg 3ul Taq 1ul ddH2O 24ul
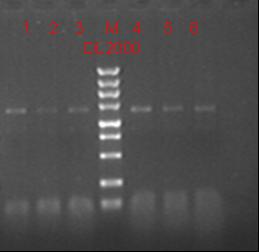
8/27/2011Phoenie: We test the co-transformed bacteria with UV system and lac system by comparing the different luciferase activity under the stimulated and non-stimulated condition. The result is confusing as usual. Zhangbo; 8.27 Binding the product together Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Nachte:Using the GFP and RFP made yesterday we got two different reporter parts which have digestion sites and homologous sequence,but it seem that primers didn't work well.False priming lead to a lot of bands in various length. To solve this problem,firstly we choose the overlapping parts who performed better as the template,and then use GFPdig-F and RFPdig-R to amplify part with digestion sites. fragment 1ul primers 1/1ul dNTPs 5ul 10*buffer 5ul Mg 3ul Taq 1ul ddH2O 33ul Finally,we saw clear bands in electrophoresis. 8/28/2011Phoenie:We discuss the possibilities that caused the abnormal data and verify our surmise by a group of preliminary trials to exame the following facts: different batches of reagents, the use of re-suspension solution and freezing. the result shows that reagents that once store at 4 ℃ can greatly effect the read-out but other facts make little different. Besides that we accidentally find out that the DNA concentration read-out can not give efficient information about the lysis of bacteria because the re-suspension solution and lysis solution give similar read-out Zhangbo8.28 Testing PCR buf 2ul dNTP 2ul Mg2+ 1.2ul F/R(WtRNA-1) 0.8/0.8ul Plasmid(result) 0.8ul KOD 0.4ul ddH2O 12ul Sequencing 8/29/2011test the 4AGG codon UV system again, with new batch of reagent: co-transformed 4AGG codon UV UV system, UV+; co-transformed 4AGG codon UV system, UV-; only 4AGG codon UV pReporter, UV+; only 4AGG codon UV pReporter, UV-; Zhangbo: 8.29 Use the plasmid from Zhaobing to construct the pACYC184-T7-RFP/RFPM 8/30/2011Zhangbo: 8.30 RFP PCR PCR KOD buf 5ul dNTP 5ul Mg2+ 3ul F/R(RFP) 2/2ul Plasmid(13M RFP) 1ul KOD 1ul ddH2O 31ul Wild left mutate right Purified Buf 2ul DNA 16ul(plasmid) Nde1 1ul Xho1 1ul Total 20ul 37℃ 5hours Nachte:EcoRI and XholI condigestion pET28a-Amp 16ul FD buffer 2ul EcoRI 1ul XholI 1ul programm:37℃*6h connection process T4 buffer 1.5ul PEG 1.5ul plasmid 2 ul 'G-L-R'fragment 4ul T4 lingase 1ul ddH2O 5ul 22℃*2h,70℃*5min 8/31/2011Zhangbo: Binding the product together Transfer the plasmid into the DH5a 10ul into 100ul DH5a 30min on the ice 37℃ 2min and 5min on the ice 220rpm 1 hour Tet solid medium for 16 hours to 24 hours Nachte:Pony did transformation last night,and I pick colonies this morning.Using colony PCR to test whether the digestion is successful. Samples A-1 A-3 T-2 T-3 T-4 T-6 T-8 are positive.After 6h's shaking,we extracted plasmid. |
 "
"