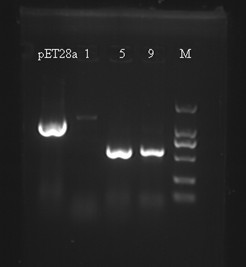
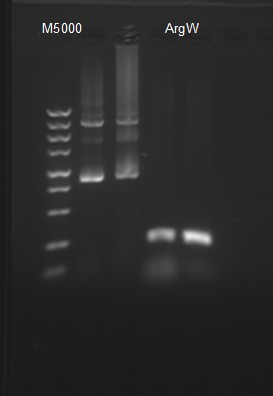
Team:SJTU-BioX-Shanghai/Notebook/Lablog
From 2011.igem.org
|
|
Calendar
July
7/15/2011Today Mr MA said we were serious about the project!! From Cathy /\ /\
{ ^ ^ }
(>:::<) //
(…:::::…)/
Phoenie: Discuss modeling with Guanhao and design the primer. And then: get locked outside my dorm until Bear finally came back from Kendo training at almost 11:00 p.m.= = (Bear: Get wounded during the training (But it is so wonderful!!). When I came back, (here I leave out some LOLs)) Nachte:Why do the above lady think my wound is wonderful=^=It drawn me both from the Kendo class and the experiment.So today my job is just learning about our vectors and target gene and practice using primer premier. From Chunying My first report got criticiasm from Mr Ma and I really needs more training to be qualified enougth to undertake my responsibilities. To give a concise presentation is not as easy as we see other do~~And I finally paid the visa fee for all the team members. zhangBO: The work would start in a few days for the primer doesn’t arrive. The overlapping PCR is a new technology to me without enough confidence leads to the anxious these days. 7/16/2011God WANG has sent me to do the gel electrophoresis~~ all 12 got the target band. From Cathy Phoenie: Waiting for the primers to be sent for a whole morning and finally give up and discuss primer designing with Bear. Hot day!⊙﹏⊙b汗 zhangBO: The primer arrives with the start of the work and the final failure of the PCR which may caused by the design of the primer. We start a new attempt of the concentration ratio. Nachte:Forma vix maken~ 1.Contacting with the Prof.Tang and the Chinese Academy Of Science to get pACYCDuet-1 and pTrc99B 2.To get the fragment as we called lacI and then insert into vector pACYC,after which we catch pTend
7/17/2011ZhangBO:The new attempt failed and I design the primer for the tRNA of the trp pairs. Nachte: Asking Prof. Enduo Wang for the plasmid,I was helped, but was also told that it's not the normal way to borrow things from other lab.
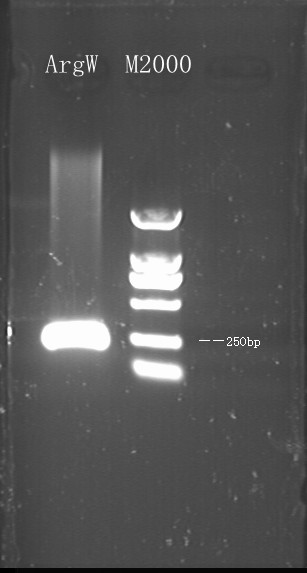
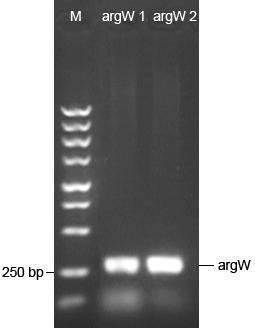
7/18/2011Phoenie: I feel a little nervous when Chunchun informed me that the problem of N-terminal of luciferase [firefly] PCR after PCR with Chunchun. and Super-You volunteered to finish the rest of experiments. We come up with the more carefully-designed method to insert target codons~ Chunying: 1.We amplified the luc gene from pGl3 basic and the luc6 strain by PCR and the result showed that the later one got more specific band.And we will later replace the Amp gene with this fragment to get the p-Reporter . 2.We amplified ArgW(tRNA gene) from E.coli's genome and replace the sulA gene on the p-Switch with it.Then WY helped us to transformed the p-Switch to E.coli.
Na chte: Ordering primers to add ArgW into the lacI fragment…Being taught overlapping PCR by God-Wang ……Being told a helpful book 'Short Protocols in Molecular Biology' which includes the sequence of cloning sites of our vectors as well as nearlly all the modern molecular biology tech.
7/19/2011Phoenie: I find that I have made a very horrible mistake: two of the primers I designed target at the wrong site. 。。。T_T。。。 Then debug. and test whether sulA operon works. Chunying: We tested the UV operon by placing the bacterial under UV for 20 seconds after cultivated it for 5 hours in about 6ml liquid culture medium. But I have made a mistake for doing this before the bacterial coming to their logarithm middle. So WY decided to cultivate it for another 3 hours and give it a second UV exposure. Tomorrow we will the product of sulA protein by SDS-PAGE. Pony: Bear and I began our expriments tonight! We transformed BioBricks BBa_I13522(GFP) and BBa_I135211(mRFP) into E.coli(DH5α) to test if the Biobricks work. Expect they will fluoresce tomorrow! Zhangbo: The attempt on the changing of the temperature failed.
7/20/2011Phoenie:Today we had a seminar and our tutor Ma Gang gave Chunchun and I some suggestion for further experiment. Pony:The BBa_I13522(GFP) was successfully transformed, it grows lovely tiny colonies~O(∩_∩)O~ But the BBa_I13522(mRFP) was failed...T_T... Zhangbo:Transfer the plasmid into the DH5a and grow on the LB and tRNA PCR. Nachte:First try of transform is successfully,but there may be something wrong with the biobrick of RFP?!
7/21/2011Phoenie: Today is such a happy and busy day. Chunchun and I have conducted 13 PCRs. Chunchun also negative-selected an E.coli culture. I extracted plasmid from 2 strains of E.coli but one of them turned out to be unusable. By the end of the day, bear told us that she had adopted a bunny in SJTU bbs and she will take it tomorrow morning. I proposed that we call it "Megi". If we read this name in reverse direction, we get "igem". Chunchun says that it sounds like "Magic" and it may bring about good luck.
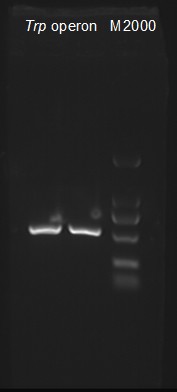
7/22/2011Phoenie: we transformed E.coli with the plasmid we have prepared yesterday and cloned the trp operon with touchdown PCR. By the way, our GRE score is available today, And the result... I guess we have done so "good" that we can settle down to experiments without any other concerning. (( /|_/| \\.._.' , ,\ /\ | '.__ v / ———— For me, GRE means God Recites English (_ . / " ) _)._ _ / @ @ '.\ \|( / ( | | ....‘‘ ’‘\\ \\....\\\|///\\\|/// Pony: Grow 4 DH5α colonies with GFP in LB tubes. Nachte:First step of building pTEnd,get ArgW from Ecoli. 's genome.When designing the primers ,I gave up terminator of ArgW itself,which is diferrent from our team-1's idea. I don't think Magi is a perfect name for a mascot of the lab, a really kick-ass name should sound valse -like ZannenJr. or Barnaby. Chunying:
7/23/2011Alfred:I did a PCR for truncated AspS gene from DH5α's genome. Pheonie: Nice day and we are very lucky in experiments~ and bunny is an interesting animal Chunying: We conducted a negative screening for the p-Reporter. Later in the day, the result came out and we inoculated the colonies that have the desired plasmid into the LB+Kana medium. We tested the result of the insertion of TrpRS and ArgW genes respectively by bacterial PCR and the electrophoresis indicated that we have successfully replaced the sulA gene with the genes we inserted 7/24/2011Phoenie: We extracted 2 plasmid and inserted trp operon into one of them by PCR, then we digested it and transformed E.coli with the product. We conducted positive test for the pET28a-luc, and got very strange result: none of the 20 picked strains is positive. We did trouble-shooting by changing the annealing temperature and test whether the backbone of the plasmid is right by PCR. Pony: The pSB1A2-GFP/DH5α grows very well; it emits bright green fluorescence. We extracted the plasmids pSB1A2-GFP from DH5α and checked it by electrophoresis . ZhangBo Design the primer for aaRS and find out that the KANA resistant mutation failed for only one colony doesn’t grow on the KANA 7/25/2011Pony: We transformed several plasmids with mRFP into DH5α again. The plasmids are pSB1A3-mRFP from 2011 and 2010 distributions(BBa_I13521) and pSB1A2-mRFP from 2011 distribution(BBa_J5526).We increase the quantity of competent cells and DNA. The DH5α competent cells was newly prepared. Alfred: I amplified the AspV gene(tRNA-Asp) from DH5α genome through PCR, then link it to pACYC plasimd with homologic arms.At last, try to transform it into DH5α competent cell, but failed:( ZhangBo: Transfer the tRNA inti the plasmid and grow on the LB 7/26/2011Alfred: I linked the AspV gene to pACYC plasmid again. After PCR, use DpnI enzyme to digest original methylated plasmid and transform pACYC-AspV to competent cell. Pony: The tansformation of DH5α with pSB1A3-mRFP was succeed! It emit red fluorescence with 6 hours. We amplified argW gene(encode tRNA-arg) from E.coli genome by PCR and checked it by electrophoresis. ZhangBo: The tRNA succeeds in growing on the LB of the right resistant. 7/27/2011Chunying: There is still something wrong with the pReporter. We have purified the PCR product before transformation, but the result didnot change for this. This night we tried to change the annealing temperature a little higher, because the low annealing temperature might result in false pairing. Reamplify ArgW gene from E.coli's genome. The plasmids are transformed into cell after they were inserted . Alfred:There are so many clonies of pACYC-AspV on the plant,so happy:) Select 8 clonies for further incubation and extract the plasmid with kit Pony: The pSB1A3-mRFP/DH5α grows very well. The excessive expression of mRFP made the bacterial suspension looks like grapefruit juice. We extracted the plasmid pSB1A3-mRFP, and the electrophoretic analysis shows it is the right plasmid. ZhangBo: Redesign of the KANA resistant mutation with the successful PCR on the aaRS. 7/28/2011Chunying: Change the annealing temperature to insert luc gene. Transform the PCR product(supposed to be pET28a-luc)into the cell. Two kinds of mutated luc gene are inserted into pET28a,and only the 'trp'mutation grow on the plate.The negative screening select one stain. Pony: Transformed the pTrc99B into competent cells DH5α.The plasmid pTrc99B was given by SIBS(Shanghai Institutes for Biological Sciences); it will be used to construct vector pTEnd. Alfred:I validated the pACYC-AspV vector through PCR and the result was nice. Nachte:Can not wait for Prof.Tang's plasmid anymore=*=We decided to plug tRNA coding sequence into the part 'LacI' at once and asked for a new pair of primers to connect 'LacI' and pACYC184. 7/29/2011Alfred:digest the TDRS and pET-28a with NdeI/NcoI enzyme in 37℃ for 8 hours, then link them with T4 ligase. Phoenie: digest PCR production with Dpn1 to purify it and transform E.coli cell and spread the cells in plate containing kana. Plan to re-clone AGG2 and wild-type luciferase but find that the KOD is used up. Nachte:Step one of the overlapping plan is smoothness,then to pay back, the last step became the Red Sea. 7/30/2011Phoenie: No enzyme come today... and I only negative select the AGG4 and AGG6 colony. but I come up with new idea about modeling Nachte:Keep fighting for final product of pTEnd.../^\ 7/31/2011Phoenie: No enzyme comes today again...get 2 positive strain of AGG6 |
 "
"