Team:Amsterdam/Notebook/Protocols/ColonyPCR
From 2011.igem.org
(Difference between revisions)
(→Protocol) |
|||
| Line 60: | Line 60: | ||
| - | |||
| - | |||
{{:Team:Amsterdam/Footer}} | {{:Team:Amsterdam/Footer}} | ||
Latest revision as of 00:01, 22 September 2011

Colony PCR
We used this protocol to check if the new transformants contained the right plasmids/insert.
Materials
- Petri dish with LB agar and appropriate antibiotic
- ON colonies on a petri dish with LB agar and appropriate antibiotic
- dNTPs
- primers (VF2 and VR)
- Taq polymerase
- Taq buffer
- dH2O
- PCR tubes
- PCR machine
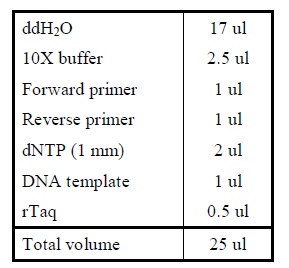
Protocol
| Sample | µl |
|---|---|
| VF2 primer | 0,5 |
| VR primer | 0,5 |
| dNTPs | 1 |
| Taq buffer | 2,5 |
| Taq polymerase | 0,2 |
| H2O | 20,3 |
| Total | 25 µl |
- Make the PCR mix and pippete it in a PCR tube.
- Pick a colony from a LB-agar plate using a sterile pipette tip.
- Pippete up and down in the PCR mix.
- Cross the pippete tip on the new LB-agar plate in the box corresponding to the sample number.
- Redo this with all the selected colonies.
- Run the following PCR program:
Note: Adjust the elongation time according to the expected product size. 1 minute for each Kb.
- Incubate the selected colonoies on the LB-agar plate ON at 37°C.
- Check the PCR-sample sizes on agarose gel.
 "
"












