Team:Amsterdam/Biobricks/Basic Parts
From 2011.igem.org

Basic Parts
Three different types of basic parts were used to construct our CryoBricks: promoters, ribosome binding sites (RBSes) and coding regions. This page contains information on their purpose in the project. Emphasis is placed on the reason why we selected these particular parts, and in case of the coding regions, the biological function of the encoded protein. For more information on the constructs into which these parts were combined, refer to the composite parts page.
Promoters
Several criteria were considered in selecting promoters for use in our CryoBricks. An ideal promoter would be: available from the parts registry, so that we can easily obtain and incorporate it; active at low temperatures, so that it works when we need it to; and positively regulated, so it won't be active unless we want it to. A search for promoters that matched one or more of these criteria resulted in the following selection:
pLacI
The Lac operon's promoter, which is often used to induce expression of proteins from plasmids artificially transformed into E. coli. Literature has numerous examples of this promoter being used to (over)express proteins at low temperatures, and it's available from the registry as [http://partsregistry.org/Part:BBa_R0010 BBa_R0010]. Unfortunately, it's not positively regulated, but its activity can be controlled to a certain extent. It's negatively regulated by the LacI protein, and as such can be stimulated by repressing LacI with IPTG or lactose. However, it will be active constitutively in strains that don't express LacI.
pBAD
The promoter of the araBAD operon. Its biological function is similar to that of pLacI; in its natural context, it activates transcription of enzymes involved with the consumption of a carbon source, when this carbon source is available to the cell. Its activity is mediated by the AraC protein, which either activates of represses expression, depending on the concentration of L-arabinose.[http://www.pnas.org/content/85/15/5444.full.pdf] The pBAD promoter is available from the registry as [http://partsregistry.org/Part:BBa_I13453 BBa_I13453], but team British Columbia 2009 derived two new promoters from this part, responding either more or less strongly to the presence of arabinose (Stronger response: [http://partsregistry.org/Part:BBa_K206000 BBa_K206000], weaker response: [http://partsregistry.org/Part:BBa_K206001 BBa_K206001]). These pBAD derived promoters were selected because of their different arabinose sensitivity. This difference can facilitate finetuning the expression levels of proteins while using a single inducer, which can be further tweaked by combining the promoters with different RBSes.
hybB
The promoter of the hydrogenase II operon. It's also known as the coldshock promoter, because it's inactive at 37°C and activates at temperatures below 30°C. Using this promoter would allow us to only have our cells express CryoBricks when E. coli actually benefits from this, but makes inducing expression in a dose-dependent manner impossible, as it's not induced by a molecule we add. It enables CryoBrick expression to be directed on 'autopilot', which has pros (for example, in large-scale industrial applications) and cons (in case CryoBricks comprising such a promoter cross over to an uncontrolled environment, be it by accident or design). The registry submission of this brick, [http://partsregistry.org/Part:BBa_J45503 BBa_J45503], never made it past the planning stage, but the brick is incorporated in several composite parts.
Final selection
All selected promoters fulfilled the availability criterion, being submitted to the registry, but hybB was only available as part of an assembled construct. The construct was ordered and preparations were made to PCR it out of this construct, but constraints on time and safety considerations caused us to decide against using it in the end.
The project was supposed to include a characterisation experiment to quantify the remaining promoters' strengths at low temperatures. Unfortunately, technical difficulties with cloning, and subsequent assembly of the reporter constructs required for this, made such characterisation infeasible. Even though we couldn't confirm pLacI and pBAD's activity at low temperatures, it was decided to construct our CryoBricks with both promoters. Of the available pBAD promoters, we selected the one that responded most strongly to arabinose ([http://partsregistry.org/Part:BBa_K206000 BBa_K206000]). It allows for the highest the maximum expression level, and any of the expression levels attainable with the related promoters can be achieved as well, given a finely tuned concentration of arabinose.
RBSes
Similar to our promoter selection criteria, the ideal RBS for our CryoBricks should be active at low temperatures, and available from the registry. We suspect temperature may have quite a drastic impact on how an RBS functions; RBSes form structures in mRNA that mediate ribosome binding, but thermodynamics govern the dynamicity (and stability) of such structures. Literature study revealed only very little information about how temperature affects the activity of RBSes. To fill this gap in the knowledgebase, we planned to characterise some of the RBSes available from the registry, using reporter constructs to measure their strength at low temperatures. Unfortunately, technical difficulties and tight deadlines impaired these plans as they did with the plans to characterise promoter strengths.
Forced to omit characterising the strength of different RBSes at low temperatures, we decided to take our chances with the RBS team Warsaw 2010 defined as their standard against which to compare other RBS strengths ([http://partsregistry.org/Part:BBa_B0034 BBa_B0034]). Running on the crude assumption low temperatures affect all RBSes in an equivalent manner, we reasoned this RBS - which is relatively strong compared to other RBSes at 'normal' temperatures - should be relatively strong at low temperatures too.
Coding regions
The novelty of the icE. coli project lies in its new submissions to the registry: 5 different coding regions, each encoding a protein reported to enhance cold resistance one way or another. Cold resistance, in this context, is defined as protein mediated mitigation of cold-induced systems failure. Such systems failure can be caused on many different levels, such as a decrease in the fluidity of the membrane; an excessive increase in structural rigidity of proteins, DNA and RNA; a drop in the rate of (bio)chemical reactions; and in extreme cases, mechanical damage caused by ice crystallisation. More often than not, low temperatures cause failure on several levels simultaneously, and mitigation of the effect on one level may be cancelled out by failure of another. In some cases, though, systems failure may be limited or prevented entirely by the expression of specific (combinations of) proteins.
Literature study reveals several proteins have been reported to decrease E. coli 's sensitivity to low temperatures. Team Amsterdam selected 5 of these proteins to create cold resistance BioBricks, or CryoBricks. They are all chaperones, originally found in cryophillic bacteria which thrive in cold environments, like the Arctic. The selected proteins are:
- Cpn10 and Cpn60, from Oleispira antarctica
- SheDnaK, from Shewanella sp. AC10
- CspA and CspC, from Polaribacter irgensii
All these proteins are, at least to a certain degree, homologous to chaperone proteins endogenous to E. coli. In addition, function loss of these endogenous chaperones is associated with cold-induced systems failure. Theoretically, expression of the cryophile chaperones listed above compensates for the loss of function endogenous chaperones experience at low temperatures, in doing so mitigating the impact of these temperatures on cellular activity.
Note that because of the many different levels at which cold-induced systems failure can occur, many different ways of characterising cold resistance are employed in different articles. Two prevalent ones used to characterise our CryoBricks are:
- the effect expression of a (combination of) protein(s) has on an organism's ability to survive freezing and thawing
- the effect expression of a (combination of) protein(s) has on an organism's specific growth rate at suboptimal temperatures
Especially the latter interpretation is of value to some of our applications. Below, you'll find information on the biology of the selected chaperones, detailling how they contribute to cold resistance in E. coli. For more information on the methods and results of characterising them, refer to the CryoBrick characterisation page.
Unfortunately, because transferring the parts to the pSB1C3 backbone proved too difficult in most cases, only [http://partsregistry.org/wiki/index.php?title=Part:BBa_K538000 BBa_K538000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K538004 BBa_K538004] could be submitted to the registry as individual parts.
Cpn10 and Cpn60
Cpn10 and Cpn60 are homologous to GroES and GroEL of E. coli, respectively. The GroEL/ES chaperone system promotes the folding and/or assembly of over 30% of E. coli's cellular proteins, is required for bacteriophage morphogenesis and has a role in protein secretion.[http://www.nature.com/nature/journal/v355/n6355/abs/355033a0.html][http://onlinelibrary.wiley.com/doi/10.1002/1521-3773%2820020402%2941:7%3C1098::AID-ANIE1098%3E3.0.CO;2-9/abstract] However, it rapidly loses its refolding activity at temperatures below 37°C.[http://www.nature.com/nbt/journal/v21/n11/pdf/nbt1103-1266b.pdf] (Figure 1b) Cpn60/10, on the other hand, functions very well at these temperatures.
Ferrer et al. identified 22 housekeeping proteins involved with E. coli 's systems failure at low temperatures. They went on to demonstrate their interactions with chaperones are key determinants of activity at these temperatures. It was shown that the Cpn60 protein from O. antarctica coprecipitates with many of the proteins found in the proteome of E. coli.[http://onlinelibrary.wiley.com/doi/10.1002/pmic.200500031/abstract] Also, by transforming E. coli with Cpn10 and Cpn60, they've enabled it to grow even at freezing point. (Figure 1a) Their findings suggest inactivation of a few cold-sensitive key proteins causes systems failure in E. coli, and that cells may be 'rescued' by reactivating these genes.
SheDnaK
DnaK is a 70 kilodalton heatshock protein involved with many different processes. It's E. coli's primary Hsp70 homolog[http://www.jbc.org/content/270/18/10412.full], and its function is mostly regulated by interactions with the smaller heatshock proteins DnaJ and GrpE. It's been debated whether or not DnaK is involved with assembling the ribosome's 30S subunit, but Maki, Southworth and Culver conclusively demonstrated this to be the case.[http://rnajournal.cshlp.org/content/9/12/1418.full] It is also essential for lambda phage propagation, playing a key role in its replication by binding the lambda P protein, disassembling the lambda P:DnaB complex.[http://www.jbc.org/content/268/7/4821.abstract]
Cold-induced inactivation of EcoDnaK, combined with the inability to refold it, is reported to be a causative agent of systems failure in E. coli at low temperatures.[http://www.nature.com/nbt/journal/v21/n11/pdf/nbt1103-1266b.pdf [5]] Yoshimune et al. isolated SheDnaK from the Antarctic Shewanella bacterium, and observed it has a much higher ATPase activity at low temperatures than EcoDnaK, which is characteristic for cold-active enzymes.[http://www.springerlink.com/content/f50n0gahppxacytb/] (Figure 2) They subsequently expressed SheDnaK in a DnaK-null mutant. This did not rescue E. coli 's growth at 43°C, nor did the strain support lambda phage propagation at 30°C, but it did allow their SheDnaK comprising E. coli to grow at temperatures as low as 15°C, which was impossible for the null-mutant. We use SheDnaK in our CryoBricks based on the speculation it may recover some of the functions EcoDnaK loses at low temperatures when they are expressed in parallel to eachother.
CspA and CspC
While exposure to low temperatures represses the expression of most proteins, cold shock protein (Csp) expression is actually induced by this. Jones and Inouye showed E. coli's CspA concentration can increase over 200-fold within 1.5 hours, when exposed to cold shock.[http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.1994.tb00359.x/pdf] CspA functions as an RNA chaperone,[http://www.jbc.org/content/272/1/196.full] capable of melting stable secundary structures in mRNA, and in doing so facilitating their translation at low temperatures.[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC207108/pdf/jbacter00084-0044.pdf][http://www.jbc.org/content/274/6/3407.full] Its nucleic acid binding properties are mediated by a β-barrel structure, which is known as the cold shock domain and is common to all cold shock proteins whose structures have been clarified. In particular, two recurring motifs - RNP1 and RNP2 - are essential for cold shock proteins' ability to bind RNA.[http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.1995.tb02431.x/abstract]
E. coli 's genome comprises a family of 9 highly similar RNA chaperones (CspA through CspI), 4 of which are cold-induced.[http://www.springerlink.com/content/jtnbcavcdpg3xey4.pdf] Deletion of one, two or three of the cold-induced chaperones does not affect cell viability at low temperatures, but a quadruple knock-out strain exhibits a cold-sensitive phenotype. Xia, Ke and Inouye not only made and characterised these knock-out strains, but went on to show the cold-sensitive phenotype can be repressed by overexpressing any of the remaining members of the Csp family, with the exception of CspD.[http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.2001.02372.x/pdf] (Overexpressing CspD did, however, increase the cold resistance of the wild-type strain.) The apparent redundancy in the functioning of E. coli 's cold shock proteins suggests that they carry out a task which is key to cell survival.
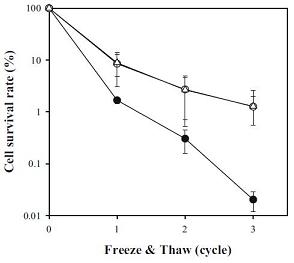
Uh et al. identified a blind spot in the scientific community's understanding of cold shock proteins, stating the insights reported above are based mainly on studies of mesophilic bacteria. To fill this gap, they investigated a bacterium isolated from Arctic sea sediment (Polaribacter irgensii) and, following a homology-based selection procedure, cloned two of its genes (CspA and CspC) into E. coli.[http://www.springerlink.com/content/qr2q5722j0t78653.pdf] They demonstrated expression of either of these genes can rescue the cold-sensitive phenotype Xia, Ke and Inouye observed in their quadruple knock-out. In addition to this, they show P. irgensii 's cold shock proteins drastically increase E. coli 's ability to survive freeze/thaw cycles. (Figure 3)
References
- Huo, Martin & Schleif Alternative DNA loops regulate the arabinose operon in Escherichia coli, Procl. Natl. Acad. Sci. USA 85, 5444-5448 (1988)
- Gething & Sambrook Protein folding in the cell, Nature 355, 33–45 (1992)
- Walter & Buchner Molecular Chaperones — Cellular Machines for Protein Folding, Angew. Chem. Int. Ed. Eng. 41, 1098–1113 (2002)
- Ferrer et al. Chaperonins govern growth of Escherichia coli at low temperatures, Nat. Biotech. 21, 1266 - 1267 (2003)
- Strocchi, Ferrer, Timmis & Golyshin Low temperature-induced systems failure in Escherichia coli: Insights from rescue by cold-adapted chaperones, Proteomics 6 (1), 193-206 (2005)
- Ziegelhoffer, Lopez-Buesa & Craig The Dissociation of ATP from hsp70 of Saccharomyces cerevisiae Is Stimulated by Both Ydj1p and Peptide Substrates, J. Bio. Chem. 270, 10412-10419 (1995)
- Maki, Southworth & Culver Demonstration of the role of the DnaK chaperone system in assembly of 30S ribosomal subunits using a purified in vitro system, RNA 9, 1418-1421 (2003)
- Osipiuk, Georgopoulos & Zylicz Initiation of lambda DNA replication - The Escherichia coli small heat shock proteins, DnaJ and GrpE, increase DnaK's affinity for the lambda P protein, J. Bio. Chem. 268, 4821-4827 (1993)
- Yoshimune et al. Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures Extremophiles 9 (2), 145-150 (2005)
- Jones & Inouye The cold shock response - a hot topic. Mol. Microbiol. 11, 811-818 (1994)
- Jiang, Hou & Inouye CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196-202 (1997)
- Jones, Krah, Tafuri & Wolffe DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J. Bacteriol. 174, 5798-5802 (1992)
- Schindler et al. The family of cold shock proteins of Bacillus subtilis - Stability and dynamics in vitro and in vivo. J. Biol. Chem. 274, 3407-3413 (1999)
- Schröder et al. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16, 699-708 (1995)
- Ermolenko & Makhatadze Bacterial cold-shock proteins. Cell. Mol. Life Sci. 59, 1902-1913 (2002)
- Xia, Ke & Inouye Acquirement of cold-sensitivity by quadruple deletion of the cspA family and its supression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40, 179-188 (2001)
- Uh et al. Rescue of a Cold-Sensitive Mutant at Low Temperatures by Cold Shock Proteins from Polaribacter irgensii KOPRI 22228 J. Microbiol. 48, 798-802 (2010)
 "
"