Team:Brown-Stanford/PowerCell/NutrientSecretion
From 2011.igem.org
CARBON SECRETION: WHY WE CHOSE SUCROSE (SILVER LAB, DUCAT CORRESPONDENCE, CELLULAR BENEFITS OF SUCROSE)
NITROGEN SECRETION: WHY WE DIDN'T FOCUS; EVIDENCE OF LEAKAGE?
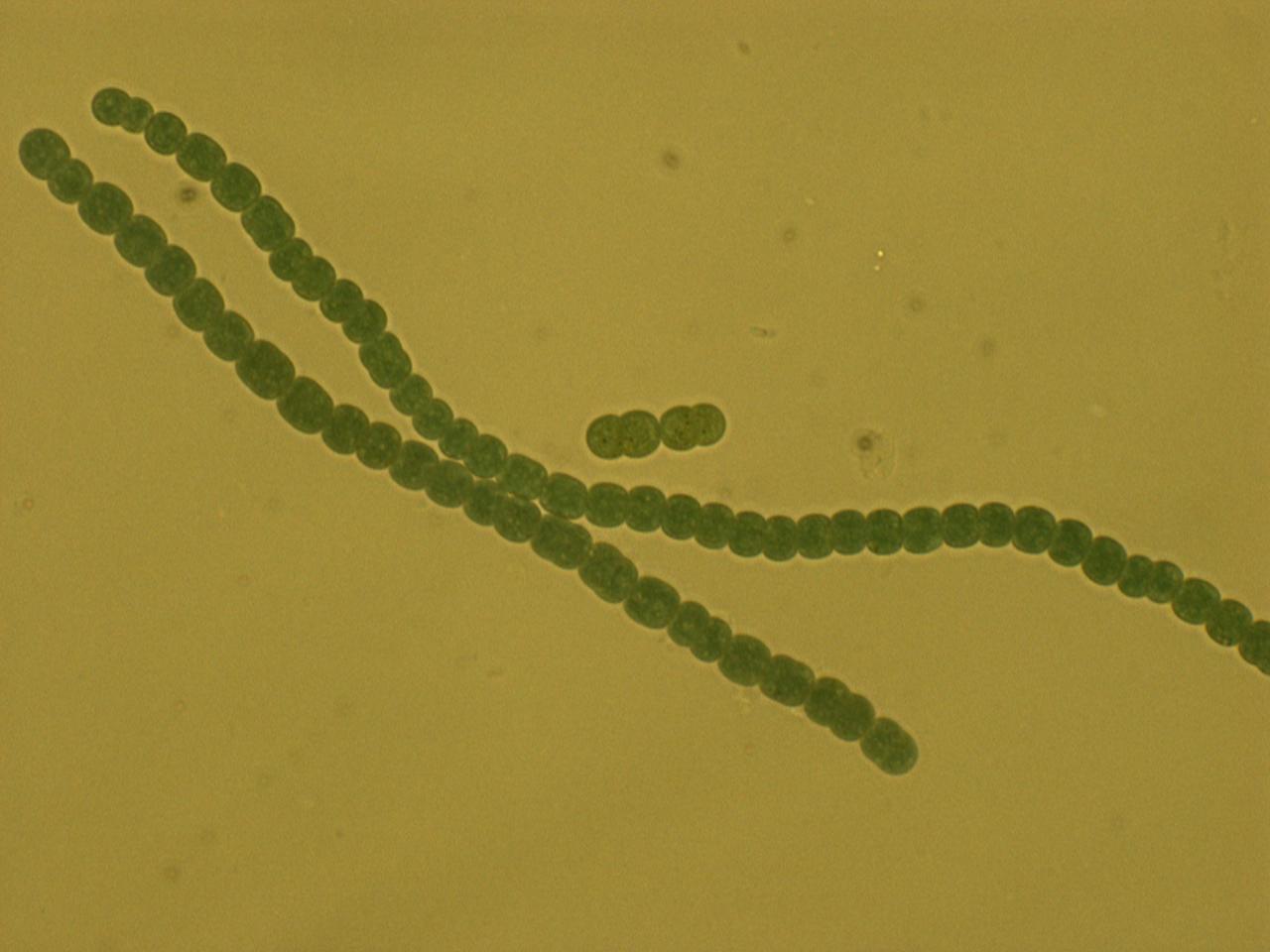
CONSTRUCT SCHEMATIC, EXPLAIN EACH COMPONENT: ANABAENA PROMOTER: HOW DID WE DERIVE THIS METHOD OF LIMITING EXPRESSION TO VEGETATIVE (CITE PAPER; SHOW PRETTY PICTURE OF LOCALIZED GFP EXPRESSION)
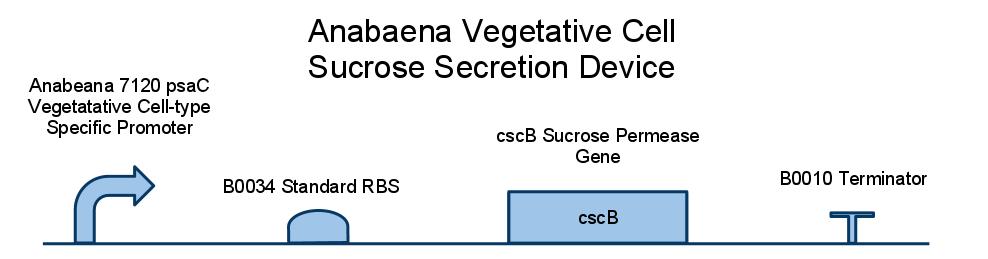
CSCB; EXPLAIN HOW SUCROSE SYMPORTER WORKS, (CITE PAPER WHERE IT IS DESCRIBED IN E COLI W)
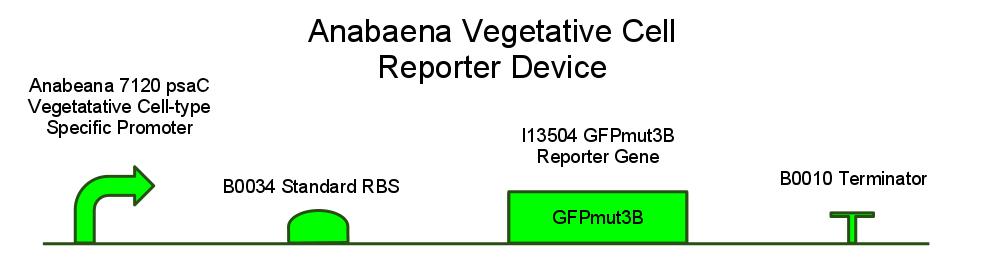
GFP REPORTER CONSTRUCT SCHEMATIC; WHAT IT IS THERE FOR, HOW WE EXPECT IT TO WORK
GFPMUT3B; WHY REGULAR GFP WON'T WORK (CITE PAPER WHERE MUT3B IS VISIBLE OVER BACKGROUND PIGMENT LEVELS)
TRANSFORMATION THIS IS HOW WE TRIED/PLAN TO GET OUR CONSTRUCT INTO OUR ANABAENA TRIPARENTAL MATING DESCRIPTION, RESTRICTION ENZYMES, HELPER PLASMIDS GALORE -MENTION CORRESPONDENCE WITH ELHAI, WOLK
Nutrient Secretion
PowerCell is the generator that will power all of the BioTools in a Martian settlement.
Like any other tools, biological tools need energy to run and raw materials to work with. The two major nutrient sources needed for biological tools are sugars (carbon sources) and nitrogenous nutrients. In a Martian settlement, PowerCell will harness the energy of the sun and matter from the surrounding atmosphere to provide both these necessities.
This summer, we tackled only the half involving sugar secretion. We built our system, however, on a platform that can support nitrogen fixation and secretion if someone develops it in the future.
For our host, we chose a diazotrophic (link to definition) cyanobacterium, Anabaena 7120. Anabaena 7120 has several special characteristics that make it an advantageous host species. First, it can fix its own nitrogen into biologically usable ammonium directly from the atmosphere, and second, it can do this at any time of day. The process of nitrogen fixation is detrimentally affected by the presence of even a small amount of oxygen. For this reason, most diazotrophs separate the processes of photosynthesis and nitrogen fixation temporally – that is, they photosynthesize in the day and fix nitrogen at night. Anabaena 7120 has a different way around this problem. It forms long filaments of cells containing two different cell types, normal (vegetative) cells, and heterocysts. Heterocysts form thick cell walls and house very anaerobic environments; they are where nitrogen fixation takes place. Normal (vegetative) cells carry on with photosynthesis. The two cell types then share nutrients up and down the filament, thus providing each cell with all the nutrients it needs.
The sugar we chose to secrete is sucrose. The major inspiration for our project was the paper [http://aem.asm.org/cgi/content/abstract/76/11/3462 Engineering Cyanobacteria To Synthesize and Export Hydrophilic Products]. This work was done in Dr. Pamela Silver's lab, and describes a glucose/fructose secretion device, achieved in the single-celled bacterium S. elongatus.
[Silver invA diagram]
In a nutshell, S. elongatus were forced under salt stress to produce sucrose, which was broken down into glucose and fructose by the glf enzyme, then transported out of the cell by the invA transporter.
At the Fifth International Conference on Synthetic Biology, SB5.0, we spoke with a member of Dr. Silver's lab, Danny Ducat. He advised us that one of the problems with the glucose/fructose secretion system is its low yield. He suspected that because glucose and fructose are directly metabolizable by the cell, much of these sugars are consumed by the cell before they ever have a chance to be secreted. For this reason, it may be better to directly secrete sucrose, which is not metabolizable by the cell, and worry about breaking it down later. This is what we did.
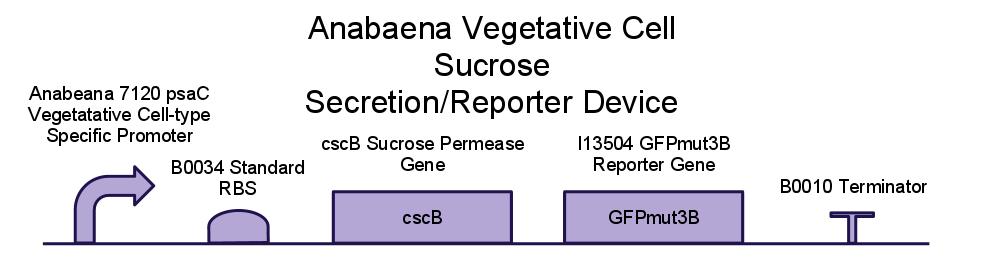
Below is a diagram of our sucrose secretion device. We placed the sucrose symporter gene, cscB, behind a promoter present in the Photo-system I of Anabaena 7120. This was done to confine sucrose secretion to only vegetative cells, as heterocysts are not producing sucrose through photosynthesis.
We have also created several other constructs containing GFP for debugging purposes. We used a modified version of GFP, GFPmut3B because normal florescence markers are easily lost among background chlorophyll pigments in cyanobacteria.
Utilizing sucrose
E. coli W
An essential aspect of the PowerCell project lies in microorganisms being able to survive off the nutrients being secreted by our genetically engineered Anabaena. Our carbon source, sucrose, is a disaccharide that is not necessarily able to be metabolized by all species.
Here our project intersects with current trends of bioproduction on Earth; sucrose is a cheap and easily obtained sugar, so there is interest in engineerable strains of microorganisms able to survive on it as a sole carbon source.
We came across research suggesting that E. coli W is one such promising strain. It is one of the few strains of non-pathogenic E. coli able to metabolize sucrose, has good tolerance for environmental stresses, and grows rapidly. (Archer 2011) There have been papers describing the process of adapting E. coli W as a bioreactor organism, and methods of growing them to high density and productivity using fed-batch cultures. (Lee 1993, Lee 1997).
In fact, the strain has already been used to produce a number of useful products. These include D(-)-lactate, a precursor to the formation of certain biodegradable plastic polymers, and L-alanine, a food additive and nutritional supplement. (Shukla 2004, Zhang 2007) With these examples, it is not difficult to imagine the production role of E. coli W in our Martian colony.
Experiment
In the application of our project, PowerCell would be grown alongside and supporting a productive microorganism strain such as E. coli W. We were thus interested in seeing whether E. coli W could grow in the conditions generated by PowerCell. This meant culturing on minimal BG-11 media with added sucrose to simulate secretion by Anabaena.
Based on literature (CITE SILVER PAPER) and correspondence with Mr. Ducat, we predicted that our Anabaena culture would produce no more than 8mM of sucrose. It would be difficult to support substantial E. coli growth on such low levels without a means of concentrating sugar via in the bioreactor design.
Initially we attempted to compare growth on BG-11 +sucrose with liquid cultures. However, multiple attempts yielded no appreciable change in O.D. after several days of incubation. We suspect that this was because E. coli growth was too small to detect via spectrophotometer.
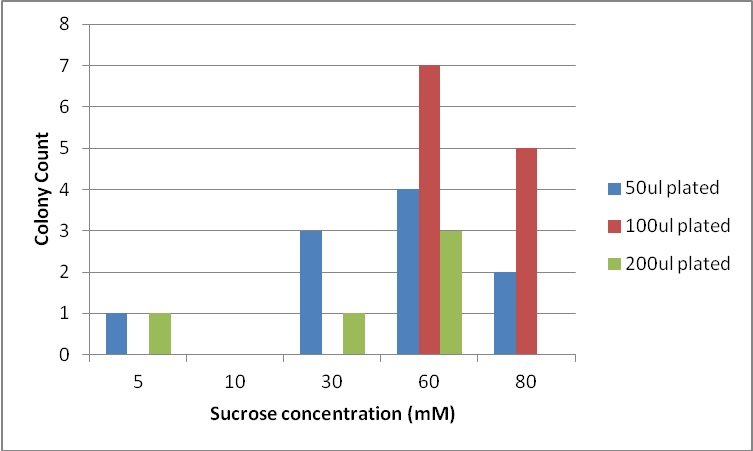
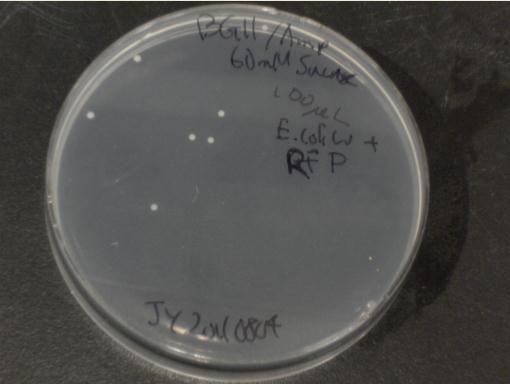
Next, we moved to agar plates as a more sensitive means of detecting cell growth. E. coli W with Amp resistance were grown, washed and resuspended in PBS to eliminate residual LB media, and streaked them on BG-11 Amp+ plates with varying concentrations of sugar (5mM, 10mM, 30mM, 60mM, and 80mM).
The cells were incubated at 37C and observed regularly before colonies appeared on the fifth day. These data suggest that, with the sugar secreted by PowerCell expressing cscB and no additional metabolic engineering, it is possible to support another organism.
References
Archer, CT et al. The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli. BMC Genomics 12:9 (2011)
Lee, SY and Ho Nam Chang. High cell density cultivation of Escherichia coli W using sucrose as a carbon source. Biotechnology Letters 15:9 (1993) 971--974.
Lee JS, Lee SY and Sunwon Park. Fed-batch culture of Escherichia coli W by exponential feeding of sucrose as a carbon source. Biotechnology Techniques 11:1 (1997) 59-62.
Shukla, VB et al. Production of D(-)-lactate from sucrose and molasses. Biotechnology Letters 26 (2004) 689-693.
Zhang, X et al. Production of L-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol 77 (2007) 355-366.
 "
"