Team:Brown-Stanford/FRETSensor/Biosensing
From 2011.igem.org
FRET and the Cellulosome
FRET
Förster resonance energy transfer ([http://en.wikipedia.org/wiki/F%C3%B6rster_resonance_energy_transfer FRET])is a phenomenon that occurs between two chromophores in close proximity (10 nm). If the emission spectra of one chromophore overlaps with the excitation of the other chromophore, then the first chromophore becomes a donor of energy to the second. This results in a greater emission of the second chromophore. This tranfer also depends on the relative orientations of the dipole moments of the donor and acceptor. FRET is commonly used as a way of visualizing biological functions, such as phosphorylation or the association of two proteins. In these cases, the target proteins are fused to fluorescent reporters such as CFP and YFP and FRET is detected during conformational changes of the proteins which close the distance between the reporters.
Atushi Miyawaki et al. created a chimeric protein using YFP, calmodulin, a glycylglycine linker, calmodulin-binding peptide M13, and CFP to detect Ca2+ binding to the calmodulin functional group. This caused a conformational change in the chimeric protein that brought the two fluorescent proteins into close proximity, causing FRET. This fusion protein increased the acceptor:donor emission ratio by 70% upon binding to Ca2+.1 Because the conformational change of this protein occurred within seconds of calcium addition, we have identified FRET as a way to produce rapid signals with GFP-based reporters as a workaround for the slow maturation times of many of its variants.
Cohesin and Dockerin
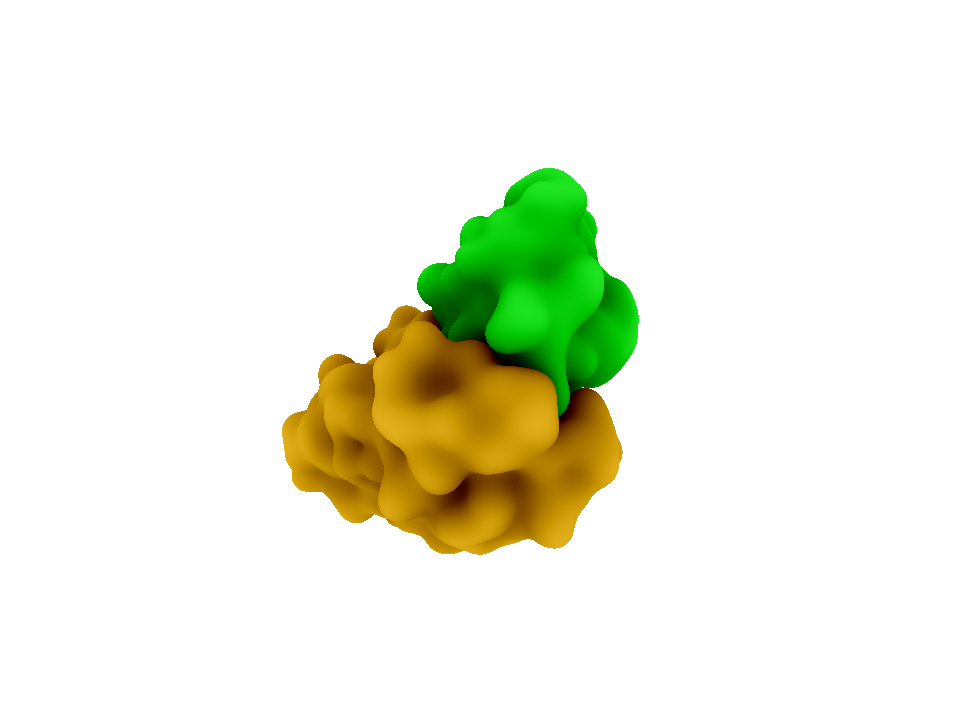
In brainstorming for a modular, highly specific method of bringing together two fluorescent proteins, we discovered the species specific interactions of the cohesin and dockerin domains of the bacterial cellulosome. Cellulose, and organic compound found the cell walls of green plants, is degraded by a protein complex called the cellulosome. The cellulosome can be found on the cell surface of many anaerobic bacteria such as [http://en.wikipedia.org/wiki/Clostridium Clostridium]. Within the cellulosome, enzyme-borne dockerin (dockerin domains on cellulases) brings the cellulose to the scaffoldin-borne cohesin, which is attached to the cell surface. These dockerin-cohesin interactions are species specific, meaning a dockerin of Clostridium cellulolyticum will not bind to a cohesin from Clostridium thermocellum2.
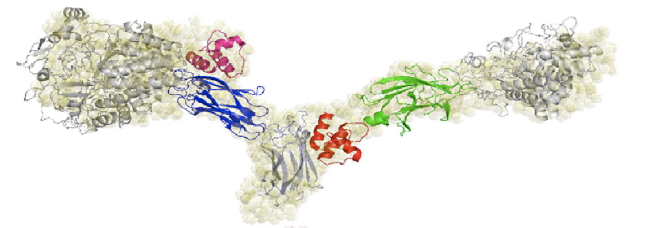
The cohesin-dockerin affinity is already being investigated as an alternative affinity tag with possible applications for protein purification3. Demishtein’s group tagged proteins with a truncated dockerin region of C. thermocellum to the C-terminus and purified the proteins using CBM-Coh and showed that the dockerin bound to a very high affinity to the CMB-Coh (cohesin). Another group of researchers, from Fierobe’s lab, has created recombinant cohesin proteins which joins one cohesin each from C. thermocellum and C. cellulolyticum with variable scaffoldiin linkers. One of their shortest linkers (Scaf4c) has 11 residues and a maximum end-to-end dimension of 98 +/-3 Å, or 9.8 nm. Because this distance is within the limit for FRET, we decided the hybrid cohesin protein can be the transcriptional product that brings together the dockerin-bound fluorescent proteins.
References
1 Miyawaki, Atsushi et al. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882-887.
2 Pages, Sandrine et al. (1997) Species-Specificity of the Cohesin-Dockein interaction between Clostridium thermocellum and Clostridium cellulolyticum: Prediction of Specificity Determinants of the Dockerin Domain. PROTEINS 29, 517-527.
3 Demishtein, Alik et al. (2010) Characterization of a dockerin-based affinity tag: application for purification of a broad variety of target proteins. Molecular Recognition 23, 525-535.
4 Molinier, Anne-Laure et al. (2011) Synergy, structure and conformational flexibility of hybrid cellulosomes displaying various inter-cohesins linkers. J. Mol. Biol. 405, 143-157.
 "
"