Team:Amsterdam/Project/Description
From 2011.igem.org

icE. coli
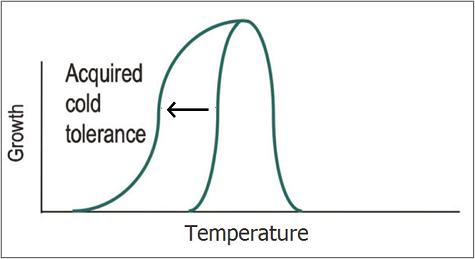
Escherichia coli's optimal growth temperature is 37°C. Its growth rate decreases drastically at lower temperatures and growth completely halts below 7°C. The aim of our project is to increase the cold tolerance of E. coli by expressing and combining several chaperone proteins that are normally found in psychrophillic bacteria. Chaperone proteins are essential in maintaining correct protein folding following changes in temperature. As such, expressing these proteins will enable enhanced growth rates at temperatures below 37°C and shift the minimal growth temperature down from 7°C, possibly even allowing growth near 0°C. Protein expression is achieved by using BioBricks, combining promoters and ribosome binding sites characterized by previous iGEM teams with newly synthesized genes that encode for psychrophile chaperones. While our subject is mainly fundamental in nature, there are various practical applications such as selection for growth on low temperatures instead of antibiotics or the optimization of biofuel production.
CryoBricks
The different proteins we intend to use for making cold resistant E. coli strains can be classified into three different categories: Cold growth rate enhancing proteins, antifreeze proteins and Freeze survivability proteins. Chaperones from psychrotrophic organisms have been show to enhance growth rate at lower than optimal temperatures. This extension of the growth curve could even shift minimum growth rate from 7°C (the wildtype minimum) to 0°C or lower. Antifreeze proteins allow for a reduction in freezing temperature as well as increased cell viability at subzero temperatures. Freeze survivability proteins allow a larger fraction of the cells to survive when being frozen and subsequently thawed. We intend to combine these three classes of proteins in order to build cells that are very well equipped against the cold. For more detailed information on any of the parts listed below, refer to our Basic Parts page in the BioBricks section.
Cold growth rate enhancing proteins
These CryoBricks are chaperone molecules closely involved with the folding of other proteins. They bind substances like RNA or other proteins, and can then inhibit or stimulate conformational changes. Such interactions may protect protection against degradation or unfolding, or even refolding of denatured proteins. This, in turn, can safeguard the functionality of a chaperone's target. Certain chaperones from psychrophillic have been been described to convey enhanced growth capabilities at low temperatures when introduced into E. coli[http://www.ncbi.nlm.nih.gov/pubmed/14595348][http://www.springerlink.com/content/f50n0gahppxacytb/]. We intend to put these genes into the BioBrick chassis and describe their effects on growth rate. Since these proteins protect our cells against cold conditions by protecting proteins from improperly folding, we suspect they might convey resistance to freezing as well. The following chaperones will be used in our CryoBricks:
- OaCpn10 (From Oleispira antarctica)
- OaCpn60 (From Oleispira antarctica)
- SheDnaK (From Shewanella sp. AC10)
Antifreeze Proteins
Antifreeze proteins, or AFPs, facilitate survival at subzero temperatures by binding small ice crystals and inhibiting their growth, preventing fatal ice crystallization. Some of them are also suspected to interact with cell membranes to protect them from cold damage. AFP has been named as superior to other substances(such as NaCl), for its ability to greatly lower the freezing temperature of water [iGEM Tokyo Tech 2009]. Other iGEM teams have also worked with AFPs, such as team Mexico 2010 and, This year, the teams of Leuven and Yale also incorporate AFPs in their respective projects. We will use their parts to allow our bacteria to live in a liquid environment at temperatures below 0°C, characterizing their growth under extreme conditions. The following AFPs will be used in our CryoBricks:
- RiAFP (From the holarctic longhorn beetle Rhagium inquisitor)
- TmAFP (From the mealworm beetle Tenebrio molitor)
- ZeAFP (From the demersal eelpout Zoarces elongatus)
Freeze survivability proteins
Freeze survivability proteins are actually cold shock proteins that allow cells to survive after being frozen. A large partition of cells still die, but a five to sixty-fold greater partition of cells has been observed to survive depending on the number of freeze-thaw cycles[http://www.ncbi.nlm.nih.gov/pubmed/21221937]. We will utilize these previously described proteins and characterize their behaviour in E. coli. Since these proteins protect our cells against cold conditions, we suspect they might enhance growth rate as well. Therefore we will also determine the effect these proteins have on growth rate in a cold environment. The following cold shock proteins will be used in our project:
- PiCspA (From Polaribacter irgensii)
- PiCspC (From Polaribacter irgensii)
Project Modules
[Under construction]
In order to successfully create a cold resistant strain of E. coli, we intend to express and combine the chaperones and AFPs listed above.
Since all action in a cell grinds to a halt at low temperatures due to reduced protein function it is essential for us to use a promoter that can effectively operate at low temperatures. Isoude.
Even though some of our targets are documented to convey increased cold growth we will still have to confirm this. Moreover, our other genes of interest may have an influence on cold growth as well and, as such, will have to be characterized in the same way.
Similarly, the effect of our targets conveying resistance to repeated freezing and thawing will have to be validated as well. For this experiment we will once again study all of our targets to see if any undocumented effects emerge.
The Project was divvied up into several project modules. Note that the first module has to be executed independently, allowing for the second and third to be executed in tandem, modules 4 and five can be started as soon as the first three modules are done. The final item can be performed based on the information gained from the fourth point.
- Characterizing and modeling promoters
- assemble a number of promoters with a strong Ribosome binding site and either a GFP or B-gal reporter
- Isoude
- Characterizing growth rate of individual Growth rate enhancing, antifreeze & freeze resistance proteins
- Use a promoter that has been shown to effectively (over?)express GFP or B-gal
- culture induced cells at 37°C and below, measuring OD600 at set intervals
- Characterizing freeze resistance proteins on cell viability
- Bas
- Combining BioBricks based on their characterization in order to maximize growth rate
- Assemble combinations of two working BioBricks into a single plasmid
- culture induced cells at 37°C and below, measuring OD600 at set intervals
- Combining Growth rate enhancing with antifreeze for subzero characterization
- Assemble combinations of two working BioBricks into a single plasmid; a growth-rate enhancer that has been shown to allow growth at 4°C with an antifreeze protein
- Make and test a cold resistance backbone
- Bas
References
- de Lorenzo Genes that move the window of viability of life: lessons from bacteria thriving at the cold extreme: mesophiles can be turned into extremophiles by substituting essential genes. Bioessays. 2011 Jan;33(1):38-42.
- Ferrer et al. Chaperonins govern growth of Escherichia coli at low temperatures. Nat. Biotechnol. 2003 Nov 21 (11):1266-7
- Yoshimune et al. Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures. EXTREMOPHILES Volume 9, Number 2, 145-150, DOI: 10.1007/s00792-004-0429-9
- Uh et al. Rescue of a cold-sensitive mutant at low temperatures by cold shock proteins from Polaribacter irgensii KOPRI 22228. J Microbiol. 2010 Dec;48(6):798-802. Epub 2011 Jan 9.
 "
"