Team:Brown-Stanford/FRETSensor/Introduction
From 2011.igem.org
(→Introduction) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Brown-Stanford/Templates/Main}} | {{:Team:Brown-Stanford/Templates/Main}} | ||
| + | <html> | ||
| + | <div id="subHeader"> | ||
| + | <ul id="subHeaderList"> | ||
| + | <li id="active"><a href="#" id="current">Introduction</a></li> | ||
| + | <li><a href="/Team:Brown-Stanford/FRETSensor/Biosensing">FRET, Cohesin, and Dockerin</a></li> | ||
| + | <li><a href="/Team:Brown-Stanford/FRETSensor/Construct">Construct</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </html> | ||
| + | {{:Team:Brown-Stanford/Templates/Content}} | ||
| - | == '''Introduction''' == | + | == '''Introduction to Biosensing''' == |
| - | + | ||
| - | + | Monitoring various aspects of the Martian environment with machinery requires a variety of probes and possibly a large amount of energy for continual detection. A more cost-effective and long-term solution to the problem of sensing changes in the environment is to use a variety of living microbes that are genetically programmed to respond to certain specific stimuli. Microbial biosensors work in essentially the same manner as a conventional sensor: the cell is equipped with a physical transducer that will induce a detectable signal once stimulated. However, the major advantage of a microbial sensor over inorganic sensors in the space environment is its ability to be mass produced from very little external input. | |
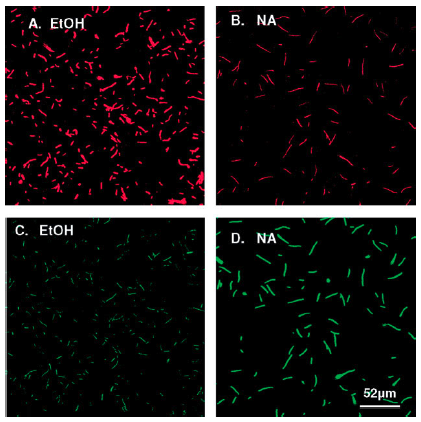
| - | + | [[File:Biosensor_Example.png|right|300px|thumb|Example of a biosensor created by Hever and Belkin induced by ethanol and nalidixic acid. From: Hever, N., Belkin, S. (2006) A Dual-Color Bacterial Reporter Strain for the Detection of Toxic and Genotoxic Effects. ''Engineering in Life Sciences'' 6.3, 319-323.]] | |
| - | + | Optical microbial biosensors come in a variety of flavors: fluorescent, bioluminescent, and colorimetric. ''In vivo'' fluorescent biosensors make use of an inducible promoter fused to a reporter gene encoding a fluorescent protein such that the protein is produced in proportion to the quantity of analyte detected. Other biosensors may also be binary and detect presence or absence. Today, biosensors, both ''in vivo'' and ''in vitro'', have myriad uses{{:Team:Brown-Stanford/Templates/FootnoteNumber|1}} and are a hot topic for iGEM teams as well. The uses for biosensors on an alien planet may be even more numerous than the uses for them on Earth. | |
| - | + | In developing a protein biosensor for DNA damage from UV radiation, we realized that one drawback is its lower sensitivity and longer response time in comparison to bioluminescent-based reporters or even biochemical reporters. On the other hand, fluorescent proteins are highly stable once formed and can be analyzed days after the detection event. These two features make reporters systems using GFP and its variants amenable to detection of trace environmental elements such as radiation dosimetry, micronutrients, toxins, other organisms, etc. Because of the longer response time, one application protein-based reporters is not suited for is a warning system. In a review of many GFP variants, the t<sub>0.5</sub> for maturation at 37°C varied from 15 minutes to over four hours{{:Team:Brown-Stanford/Templates/FootnoteNumber|2}}. | |
| + | |||
| + | Our team wanted to be able to detect rapid changes in environment or condition of the cell using fluorescent proteins and increase the scope of microbial biosensors to that of early warning systems. Ideally, a batch of cells sent to Mars should be able to alert the astronauts of DNA damage and loss of functionality due to UV radiation. Therefore, we decided to make novel use of an existing mechanism, FRET, that can make use of fluorescent proteins to image fast changes in cell state. | ||
| + | |||
| + | ==='''Thanks to'''=== | ||
| + | The FRETcetera team would like to thank [http://syntheticbiology.arc.nasa.gov/node/2#Dr._Chad_Paavola Dr. Chad Paavola], who makes beautiful multiemzyme structures from cellulosome parts, for his great ideas and constant help. We'd also like to thank Suzanne Chan, in Dr. Paavola's lab, for helping us with the protocols. | ||
===References=== | ===References=== | ||
| - | {{:Team:Brown-Stanford/Templates/Footnote|1|Su, Liang et al. Microbial Biosensors: A Review. Biosensors and Bioelectronics 26 (2011) 1788–1799.}} | + | {{:Team:Brown-Stanford/Templates/Footnote|1|Su, Liang et al. Microbial Biosensors: A Review. ''Biosensors and Bioelectronics'' 26 (2011) 1788–1799.}} |
| - | {{:Team:Brown-Stanford/Templates/Footnote|2|Shaner, Nathan et al. A guide to choosing fluorescent proteins.Nature Methods 2.12 (2005) 905-909.}} | + | {{:Team:Brown-Stanford/Templates/Footnote|2|Shaner, Nathan et al. A guide to choosing fluorescent proteins. ''Nature Methods'' 2.12 (2005) 905-909.}} |
{{:Team:Brown-Stanford/Templates/Foot}} | {{:Team:Brown-Stanford/Templates/Foot}} | ||
Latest revision as of 01:24, 29 September 2011
Introduction to Biosensing
Monitoring various aspects of the Martian environment with machinery requires a variety of probes and possibly a large amount of energy for continual detection. A more cost-effective and long-term solution to the problem of sensing changes in the environment is to use a variety of living microbes that are genetically programmed to respond to certain specific stimuli. Microbial biosensors work in essentially the same manner as a conventional sensor: the cell is equipped with a physical transducer that will induce a detectable signal once stimulated. However, the major advantage of a microbial sensor over inorganic sensors in the space environment is its ability to be mass produced from very little external input.
Optical microbial biosensors come in a variety of flavors: fluorescent, bioluminescent, and colorimetric. In vivo fluorescent biosensors make use of an inducible promoter fused to a reporter gene encoding a fluorescent protein such that the protein is produced in proportion to the quantity of analyte detected. Other biosensors may also be binary and detect presence or absence. Today, biosensors, both in vivo and in vitro, have myriad uses1 and are a hot topic for iGEM teams as well. The uses for biosensors on an alien planet may be even more numerous than the uses for them on Earth.
In developing a protein biosensor for DNA damage from UV radiation, we realized that one drawback is its lower sensitivity and longer response time in comparison to bioluminescent-based reporters or even biochemical reporters. On the other hand, fluorescent proteins are highly stable once formed and can be analyzed days after the detection event. These two features make reporters systems using GFP and its variants amenable to detection of trace environmental elements such as radiation dosimetry, micronutrients, toxins, other organisms, etc. Because of the longer response time, one application protein-based reporters is not suited for is a warning system. In a review of many GFP variants, the t0.5 for maturation at 37°C varied from 15 minutes to over four hours2.
Our team wanted to be able to detect rapid changes in environment or condition of the cell using fluorescent proteins and increase the scope of microbial biosensors to that of early warning systems. Ideally, a batch of cells sent to Mars should be able to alert the astronauts of DNA damage and loss of functionality due to UV radiation. Therefore, we decided to make novel use of an existing mechanism, FRET, that can make use of fluorescent proteins to image fast changes in cell state.
Thanks to
The FRETcetera team would like to thank [http://syntheticbiology.arc.nasa.gov/node/2#Dr._Chad_Paavola Dr. Chad Paavola], who makes beautiful multiemzyme structures from cellulosome parts, for his great ideas and constant help. We'd also like to thank Suzanne Chan, in Dr. Paavola's lab, for helping us with the protocols.
References
1 Su, Liang et al. Microbial Biosensors: A Review. Biosensors and Bioelectronics 26 (2011) 1788–1799.
2 Shaner, Nathan et al. A guide to choosing fluorescent proteins. Nature Methods 2.12 (2005) 905-909.
 "
"