Team:British Columbia/Notebook/Week 7
From 2011.igem.org
(→July 22 2011) |
|||
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Notebook}} | {{Template:Notebook}} | ||
| - | ==July 18 | + | <html> |
| - | + | <style> | |
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b>Week 7: July 17-23</b> <html><a name="w7"></a></html> | ||
| + | ==Lab Meeting - July 18== | ||
| + | Still figuring out accommodation and flight options.. | ||
| + | The team needs to continue fundraising! | ||
| - | + | Repeated sequencing at NAPS has been failing. The grad advisors (Alina and Rafael) are helping to troubleshoot. Is it NAPS? Are our samples contaminated? Rafael usually sends his samples to a sequencing company in Alabama; he suggests we send a few of our samples there first as they are supposed to be quite reliable. | |
| - | == | + | ==Human Practices== |
| - | < | + | <html><a title='By Iwona Erskine-Kellie (Flickr) [CC-BY-2.0 (www.creativecommons.org/licenses/by/2.0)], via Wikimedia Commons' href='http://commons.wikimedia.org/wiki/File:Science_World_Sunset.jpg'><img height='200' alt='Science World Sunset' src='http://upload.wikimedia.org/wikipedia/commons/thumb/a/a0/Science_World_Sunset.jpg/800px-Science_World_Sunset.jpg'/></a></html> |
| - | + | ||
| - | + | Laura was very excited to meet with Dr. Anderson on Monday to discuss our teams involvement in the Future Science Leaders program at Science World! Science World British Columbia is a non-profit organization which engages British Columbians in science and inspires future science and technology leadership throughout our province. The Future Science Leaders is a program offered to high school students who are keen and bright and want to further their understanding of science. Through weekly meetings the program will cover several difference sciences such as biology, mathematics, technology, physical sciences, earth and space, and technology. We as an iGEM team have the privilege of presenting our project to this group of high school students and their parents. We will also be educating the students on the diverse applications of synthetic biology and give a brief tutorial about what synthetic biology is! There is also an opportunity to promote the idea of a high school iGEM team for next year. | |
| - | + | ||
| - | + | ==3-Carene== | |
| - | + | [[File:Marijacob1.JPG | thumb | right | 200px | Jacob working away]] | |
| + | Daisy has been trying to put the his-tagged truncated 3-carene synthase into the yeast plasmid, PA415GPD. This has been more difficult than anticipated. The primers used to PCR out the truncated 3-carene synthase seem to produce a smear around 1.7 kb. This is the correct size range for the truncated synthase but it is not a clear band. Daisy may need to adjust the annealing temperature of her primers to get a specific band. At this point, she is going to digest and ligate this non-specific band into the yeast plasmid. She will try purification methods of gel extraction and PCR purification... | ||
| - | |||
| - | + | ==Characterization: Limonene Synthase== | |
| - | + | Daisy has decided to characterize the limonene synthase and the IDI1 (with Jacob) to fulfill the gold medal criteria. A couple of important points: | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -Limonene synthase is from '''lemon''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -Limonene synthase is a composite part (ie. Has other genes that supposedly increase terpenoid biosynthesis) | |
| - | + | ||
| - | + | -Limonene synthase is on an ampicillin resistant plasmid | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -We are not sure how well the synthase will grow in bacteria because the synthase comes from plants | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | -We plan to incorporate '''pRARE2''' plasmid into the bacteria also. The pRARE2 has rare codons that allow the bacteria to express synthases. This is from Bohlmann Lab (Chris Keeling - Thanks!) | ||
| + | -We plan to add the '''IDI1''' part from the registry into the bacteria also | ||
| - | ''' | + | -We plan to improve the '''IDI1''' part by removing a cut site (done by Jacob) |
| - | + | -IDI1 gene supposedly increases terpenoid biosynthesis | |
| - | + | ==1,8-Cineole== | |
| - | + | Jacob's sequencing also came back null. However, the restriction digest gels clearly showed that there were no cut sites in the synthase, so Jacob decided to try getting the synthases into biobrick plasmids. Three PCR attempts later... | |
| - | ' | + | |
| - | + | ==beta-Pinene== | |
| - | + | [[File:Marijacob2.JPG | thumb | left | 150px |Marianne and Jacob share their frustrations]] | |
| - | + | ||
| - | + | ||
| - | + | Marianne's sequencing attempts failed. Very frustrated at this point, she decided that she doesn't want to waste any more time on sequencing. So she restriction digested the mini-prepped plasmids to see if the site is still there. Gel verification of the digests suggested that the site has been removed (the gel showed bands identical to the uncut plasmid). | |
| - | + | Marianne PCRed off the original and the SDMed synthase using yeast primers (containing XhoI and SpeI restriction enzyme sites) in order to put it in the yeast plasmid PA415GPD. Marianne also PCRed off the SDMed synthase using biobrick primers (containing EcoRI, XbaI, SpeI, PstI restriction enzyme sites) to put it in the psb1C3 backbone. Gel verification for both PCRs shows bands at the correct length! | |
| - | + | Marianne started to assemble the biobrick part. She digested the linearized psb1C3 and the synthase gene with EcoRI and PstI using Biobrick Restriction Digest protocol. She then ligated the backbone and the part and transformed it into DH5alpha competent cells. Colony PCR of transformants using G1004 and G1005 primers resulted in no bands on the gel... Therefore, she repeated the PCR using a new batch of primers as well as a different primer pair VF2/VR. | |
| - | + | <br><br> | |
| - | + | ||
| - | + | Marianne also restriction digested the yeast plasmids and synthase genes and ligated the following 8 combinations: | |
| - | ** | + | *original + his-tagged + GAL |
| - | * | + | *original + his-tagged + GDP |
| + | *original + no-his + GAL | ||
| + | *original + no-his + GDP | ||
| + | *SDM + his-tagged + GAL | ||
| + | *SDM + his-tagged + GDP | ||
| + | *SDM + no-his + GAL | ||
| + | *SDM + no-his + GDP | ||
| + | Marianne then transformed the ligated samples. Marianne had a very long day... | ||
| - | + | ==(-)-Limonene== | |
| - | + | While the SDMs were failing, Vicki decided to try to be efficient by running another experiment in parallel: She decided to use the yeast primers to get the original (un-SDMed) synthase out, and into a yeast plasmid. However, PCR of the synthase using yeast primers resulted in 4 bands on all the lanes the gel (unexpected results! Vicki should expect a band at around 2kb). | |
| + | *Troubleshooting: use a temperature gradient from 64oC to 72oC... Result: 4 bands again. | ||
| + | *More troubleshooting: decrease the extension time to 30sec (15sec/1kb) with an annealing temperature of 70C. Vicki will also transform original plasmid (pADM743) into DH5alpha cells in case the plasmid she is working with has been contaminated somehow, and hence not being PCRed off properly. | ||
| - | + | On the other note, Vicki decided to get new primers (higher temperature and more base pairs to anneal to the template) to re-attempt the SDM... but once again, there were no bands. Vicki is extremely frustrated at this point, and is starting to doubt the integrity of the original tube (pADM743) of (-)-limonene synthase. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==ERG20 & erg20-2== |
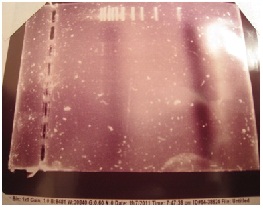
| - | + | Since they still hadn't received the plasmids from France, Gurpal decided to try generating the erg20-2 mutant by SDM. After ordering and receiving primers, his goal was to isolate the ERG20 gene from wild type yeast. Three tubes were prepared for the PCR, each containing a different amount of yeast genome, and the PCR was performed. Afterwards, Jacob ran gel electrophoresis to see if the PCR worked. However, the gel showed nothing but the ladder... just for kicks, here the is the proof: | |
| - | + | ||
| - | + | [[File: Gel_-_attempt_to_remove_ERG20.jpg| frame | center | ]] | |
| - | + | ||
| - | + | ||
| - | + | Fortunately, the ERG20 and erg20-2 genes arrived from France on pNEV-N plasmids the next day! This increased the entire team's morale 10-fold! The ERG20 gene was on the pBS plasmid and the erg20-2 was on the pKS plasmid. Gurpal successfully transformed these plasmids into <i>E. coli</i> on his second attempt using newly made Ampicillin agar plates. | |
| + | In order to replicate more cells, Gurpal made overnight cultures for both the pBS and pKS plasmids in E. coli. The following day, I had performed mini plasmid preps (using the Invitrogen kit) and had one sample of pBS plasmid and 2 samples of pKS plasmids. He used the nanodrop and determined that the concentrations were as follows: | ||
| + | [pBS] = 100.5 ng/µL | ||
| + | [pKS] = 90.0 ng/µL (first sample, labeled pKS1) | ||
| + | [pKS] = 69.2 ng/µL (second sample, labeled pKS2) | ||
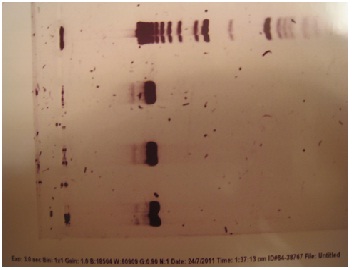
| - | + | Gurpal verified that these were in fact the correct genes. Each gene should have been about 7000 bps. After performing gel electrophoresis, he verified that his transformations and mini plasmid preps were a success! | |
| + | Here is the gel image: | ||
| - | + | [[File:Gel_-_verification_of_pBS_pKS_in_ecoli.jpg | frame | center | ]] | |
| - | + | ==alpha-Pinene== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Joe is taking a break from lab work because he has a poster day presentation to prepare for at his other summer research lab at the Child and Family Research Institute (CFRI). In the mean time, he has started looking at the arcGIS data from the B.C forestry on the pine-beetle spread while learning how to use the arcGIS software. He is also waiting for primers to come so that he can introduce cut-sites to his alpha-pinene synthase gene for PCR, in preparation for insertion into our chosen yeast constructs <html><b><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K517000"> GAL promoter BBa_k517000 </b></a></html> and <html><b><a href="http://partsregistry.org/wiki/index.php?title=Part:BBa_K517001"> GPD promoter BBa_k517001 </b></a></html> | |
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 23:12, 16 October 2011

 |
 |
 |
 |
 |
Contents |
Lab Meeting - July 18
Still figuring out accommodation and flight options.. The team needs to continue fundraising!
Repeated sequencing at NAPS has been failing. The grad advisors (Alina and Rafael) are helping to troubleshoot. Is it NAPS? Are our samples contaminated? Rafael usually sends his samples to a sequencing company in Alabama; he suggests we send a few of our samples there first as they are supposed to be quite reliable.
Human Practices
Laura was very excited to meet with Dr. Anderson on Monday to discuss our teams involvement in the Future Science Leaders program at Science World! Science World British Columbia is a non-profit organization which engages British Columbians in science and inspires future science and technology leadership throughout our province. The Future Science Leaders is a program offered to high school students who are keen and bright and want to further their understanding of science. Through weekly meetings the program will cover several difference sciences such as biology, mathematics, technology, physical sciences, earth and space, and technology. We as an iGEM team have the privilege of presenting our project to this group of high school students and their parents. We will also be educating the students on the diverse applications of synthetic biology and give a brief tutorial about what synthetic biology is! There is also an opportunity to promote the idea of a high school iGEM team for next year.
3-Carene
Daisy has been trying to put the his-tagged truncated 3-carene synthase into the yeast plasmid, PA415GPD. This has been more difficult than anticipated. The primers used to PCR out the truncated 3-carene synthase seem to produce a smear around 1.7 kb. This is the correct size range for the truncated synthase but it is not a clear band. Daisy may need to adjust the annealing temperature of her primers to get a specific band. At this point, she is going to digest and ligate this non-specific band into the yeast plasmid. She will try purification methods of gel extraction and PCR purification...
Characterization: Limonene Synthase
Daisy has decided to characterize the limonene synthase and the IDI1 (with Jacob) to fulfill the gold medal criteria. A couple of important points:
-Limonene synthase is from lemon
-Limonene synthase is a composite part (ie. Has other genes that supposedly increase terpenoid biosynthesis)
-Limonene synthase is on an ampicillin resistant plasmid
-We are not sure how well the synthase will grow in bacteria because the synthase comes from plants
-We plan to incorporate pRARE2 plasmid into the bacteria also. The pRARE2 has rare codons that allow the bacteria to express synthases. This is from Bohlmann Lab (Chris Keeling - Thanks!)
-We plan to add the IDI1 part from the registry into the bacteria also
-We plan to improve the IDI1 part by removing a cut site (done by Jacob)
-IDI1 gene supposedly increases terpenoid biosynthesis
1,8-Cineole
Jacob's sequencing also came back null. However, the restriction digest gels clearly showed that there were no cut sites in the synthase, so Jacob decided to try getting the synthases into biobrick plasmids. Three PCR attempts later...
beta-Pinene
Marianne's sequencing attempts failed. Very frustrated at this point, she decided that she doesn't want to waste any more time on sequencing. So she restriction digested the mini-prepped plasmids to see if the site is still there. Gel verification of the digests suggested that the site has been removed (the gel showed bands identical to the uncut plasmid).
Marianne PCRed off the original and the SDMed synthase using yeast primers (containing XhoI and SpeI restriction enzyme sites) in order to put it in the yeast plasmid PA415GPD. Marianne also PCRed off the SDMed synthase using biobrick primers (containing EcoRI, XbaI, SpeI, PstI restriction enzyme sites) to put it in the psb1C3 backbone. Gel verification for both PCRs shows bands at the correct length!
Marianne started to assemble the biobrick part. She digested the linearized psb1C3 and the synthase gene with EcoRI and PstI using Biobrick Restriction Digest protocol. She then ligated the backbone and the part and transformed it into DH5alpha competent cells. Colony PCR of transformants using G1004 and G1005 primers resulted in no bands on the gel... Therefore, she repeated the PCR using a new batch of primers as well as a different primer pair VF2/VR.
Marianne also restriction digested the yeast plasmids and synthase genes and ligated the following 8 combinations:
- original + his-tagged + GAL
- original + his-tagged + GDP
- original + no-his + GAL
- original + no-his + GDP
- SDM + his-tagged + GAL
- SDM + his-tagged + GDP
- SDM + no-his + GAL
- SDM + no-his + GDP
Marianne then transformed the ligated samples. Marianne had a very long day...
(-)-Limonene
While the SDMs were failing, Vicki decided to try to be efficient by running another experiment in parallel: She decided to use the yeast primers to get the original (un-SDMed) synthase out, and into a yeast plasmid. However, PCR of the synthase using yeast primers resulted in 4 bands on all the lanes the gel (unexpected results! Vicki should expect a band at around 2kb).
- Troubleshooting: use a temperature gradient from 64oC to 72oC... Result: 4 bands again.
- More troubleshooting: decrease the extension time to 30sec (15sec/1kb) with an annealing temperature of 70C. Vicki will also transform original plasmid (pADM743) into DH5alpha cells in case the plasmid she is working with has been contaminated somehow, and hence not being PCRed off properly.
On the other note, Vicki decided to get new primers (higher temperature and more base pairs to anneal to the template) to re-attempt the SDM... but once again, there were no bands. Vicki is extremely frustrated at this point, and is starting to doubt the integrity of the original tube (pADM743) of (-)-limonene synthase.
ERG20 & erg20-2
Since they still hadn't received the plasmids from France, Gurpal decided to try generating the erg20-2 mutant by SDM. After ordering and receiving primers, his goal was to isolate the ERG20 gene from wild type yeast. Three tubes were prepared for the PCR, each containing a different amount of yeast genome, and the PCR was performed. Afterwards, Jacob ran gel electrophoresis to see if the PCR worked. However, the gel showed nothing but the ladder... just for kicks, here the is the proof:
Fortunately, the ERG20 and erg20-2 genes arrived from France on pNEV-N plasmids the next day! This increased the entire team's morale 10-fold! The ERG20 gene was on the pBS plasmid and the erg20-2 was on the pKS plasmid. Gurpal successfully transformed these plasmids into E. coli on his second attempt using newly made Ampicillin agar plates.
In order to replicate more cells, Gurpal made overnight cultures for both the pBS and pKS plasmids in E. coli. The following day, I had performed mini plasmid preps (using the Invitrogen kit) and had one sample of pBS plasmid and 2 samples of pKS plasmids. He used the nanodrop and determined that the concentrations were as follows: [pBS] = 100.5 ng/µL [pKS] = 90.0 ng/µL (first sample, labeled pKS1) [pKS] = 69.2 ng/µL (second sample, labeled pKS2)
Gurpal verified that these were in fact the correct genes. Each gene should have been about 7000 bps. After performing gel electrophoresis, he verified that his transformations and mini plasmid preps were a success! Here is the gel image:
alpha-Pinene
Joe is taking a break from lab work because he has a poster day presentation to prepare for at his other summer research lab at the Child and Family Research Institute (CFRI). In the mean time, he has started looking at the arcGIS data from the B.C forestry on the pine-beetle spread while learning how to use the arcGIS software. He is also waiting for primers to come so that he can introduce cut-sites to his alpha-pinene synthase gene for PCR, in preparation for insertion into our chosen yeast constructs GAL promoter BBa_k517000 and GPD promoter BBa_k517001
 "
"