Team:British Columbia/Notebook/Week 4
From 2011.igem.org
Watermelon (Talk | contribs) |
|||
| (35 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Notebook}} | {{Template:Notebook}} | ||
| + | <html> | ||
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b>Week 4: June 26-July 2 </b> <html><a name="w4"></a></html> | ||
| - | + | ==Lab Meeting - June 27== | |
| - | [[File:ubcnotebook.jpg]] | + | [[File:ubcnotebook.jpg | frame | right ]][[File:ubcwk5pcrtubes.jpg| thumb | right | 200px]] |
We created a set of lab rules due to common mistakes made in the lab over the past few weeks. | We created a set of lab rules due to common mistakes made in the lab over the past few weeks. | ||
| Line 13: | Line 17: | ||
For our modeling projects, Jacob obtained the ArcGIS program for modeling the pine beetle infestation and Gurpal created a wet lab flow chart to match the modeling. | For our modeling projects, Jacob obtained the ArcGIS program for modeling the pine beetle infestation and Gurpal created a wet lab flow chart to match the modeling. | ||
| - | + | ==3-Carene== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Daisy made primers for sequencing because the mutagen site is in the middle of the gene and her flanking primers will only sequence 700-800bp into the sequence. | Daisy made primers for sequencing because the mutagen site is in the middle of the gene and her flanking primers will only sequence 700-800bp into the sequence. | ||
| Line 27: | Line 27: | ||
Below is an example of the chromatogram that came. | Below is an example of the chromatogram that came. | ||
| + | [[File:Sequencingseq.png | frame | left]] | ||
| - | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> | |
| + | ==1,8-Cineole== | ||
| - | |||
| - | [[File: | + | [[File:ubccookglove.jpg | thumb | right | 300px]] |
| + | Gurpal attempted to remove restriction sites from the PgxeTPS-Cin synthase gene, for Jacob, by doing a SDM-PCR. After doing it, he ran the PCR product through a gel and nothing showed up. Fortunately... | ||
| + | |||
Jacob ran successful a SDM-PCR to remove a restriction site as verified by restriction digest and gel electrophoresis. | Jacob ran successful a SDM-PCR to remove a restriction site as verified by restriction digest and gel electrophoresis. | ||
| - | He transformed competent cells with his SDM-PCR product and also ran SDM-PCR on | + | He transformed competent cells with his SDM-PCR product and also ran SDM-PCR on another, closely related synthase (isolated from a hybrid between Picea glauca and Picea englmanii, as opposed to a pure Picea glauca). He ran Raf's mini-prep protocol on transformed cells to isolate a plasmid containing the SDMed pg-TPS-cin gene. |
| + | |||
| + | ==alpha-Pinene== | ||
| + | |||
| + | New Pfu polymerases were used for Joe's SDM-PCR and it worked (using the <html><b><a href="http://kirschner.med.harvard.edu/files/protocols/Stratagene_quickchangepdf.pdf"> Quikchange Site-directed Mutagensis kit </b></a></html>)! Hurrah! | ||
| + | |||
| + | He found an optimal Quikchange PCR reaction mixture (for 50uL reaction): | ||
| + | |||
| + | <table border="3"> | ||
| + | <tr> | ||
| + | <th>Reagents | ||
| + | <th>Volume | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>ddH2O</td> | ||
| + | <td>40uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10X Rxn Buffer</td> | ||
| + | <td> 5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTP (25mM</td> | ||
| + | <td> 1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Ps-TPS-Pin Fw(125ng)</td> | ||
| + | <td> 1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Ps-TPS-Pin Re(125ng)</td> | ||
| + | <td> 1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Pfu polymerase</td> | ||
| + | <td> 1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>DNA template (50ng/uL)</td> | ||
| + | <td> 1uL</td> | ||
| + | </table> | ||
| + | |||
| + | |||
| + | Cycling Conditions: | ||
| + | <table border="3"> | ||
| + | <tr> | ||
| + | <th>Cycles | ||
| + | <th>Temperature | ||
| + | <th>Time | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>95C</td> | ||
| + | <td>1 min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>16</td> | ||
| + | <td>95C</td> | ||
| + | <td>1 min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>55C</td> | ||
| + | <td>1 min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>68C</td> | ||
| + | <td>10 min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1</td> | ||
| + | <td>4C</td> | ||
| + | <td>hold</td> | ||
| + | |||
| + | </table> | ||
| + | |||
| + | Before he moved on to the DpnI digest, Joe did a gel electrophoresis on the SDMed-PCR product. The band on the gel showed up as predicted (a pet200 plasmid at around 8.6kb). Joe then did a DpnI digest SDM-PCR products with 0.5uL of DpnI and incubated at 37C overnight. Afterwards, he transformed 2uL of DpnI products on Kanamycin plates. He also did a negative control to test the plates for kanamycin. Unfortunately, there is no growth of colonies in all plates. His next step is to increase the amount of DpnI digested products in the transformation. | ||
| + | |||
| + | At first, Joe didn't bother to do a gel electrophoresis on his PCR products after DpnI digest. He later did and found all his PCR products digest for some strange reason, which makes him wonder if he only had his original DNA template. | ||
| + | His next step is to redo his SDM-PCR. So forget about the horray at the beginnning! | ||
| + | |||
| + | ==beta-Pinene & (-)-Limonene== | ||
| + | |||
| + | Marianne and Vicki ran SDM-PCR on their synthase genes and verified their PCR products through gel electrophoresis. No bands were present in any of the lanes. | ||
| + | *Troubleshooting: The dNTP could have been too old/contaminated (we had trouble with this particular tube of dNTP from last year), the concentrations for the reagents were not optimized so... Marianne looked into some protocols and optimized the SDM-PCR protocol. Let the failures begin. | ||
| + | |||
| + | In a group, Vicki, Marianne, Gurpal and Jacob again attempted to perform SDM to remove an internal cut-site. ThermoPol was used as a buffer and PFU was used. After adding the primers and other constituents, we each ended up with ~ 25uL samples. We set our samples to run in a thermocycler for about 4 hours with specified conditions. When the cycling was completed, DpnI enzyme was added to each PCR tube and incubated overnight at 37oC. | ||
| - | + | To check if our SDM worked, we performed gel electrophoresis. Unfortunately, our gel showed that our site directed mutagenesis did not work as our product was still cut in two by the restriction enzyme. But, each of us learnt what not to do for next time which is always good. | |
| - | |||
| - | |||
| - | + | <html> | |
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 23:11, 16 October 2011

 |
 |
 |
 |
 |
Contents |
Lab Meeting - June 27
We created a set of lab rules due to common mistakes made in the lab over the past few weeks.
Vicki and Marianne will have training sessions throughout the week to show team members without wet lab experience how to PCR, cast and run a gel, digest, ligate, transform, design primers and send mini-prepped products to be sequenced.
We also received and fulfilled a request from Grinnell College (a new iGEM team this year!) for the DspB part created by the 2010 UBC iGEM team.
For our modeling projects, Jacob obtained the ArcGIS program for modeling the pine beetle infestation and Gurpal created a wet lab flow chart to match the modeling.
3-Carene
Daisy made primers for sequencing because the mutagen site is in the middle of the gene and her flanking primers will only sequence 700-800bp into the sequence.
She did a single digest with EcoRI-HF to test if the EcoRI site is in the synthase or not. Chris Keeling emailed Daisy the sequence he used for the truncation of the 3-Carene synthase.
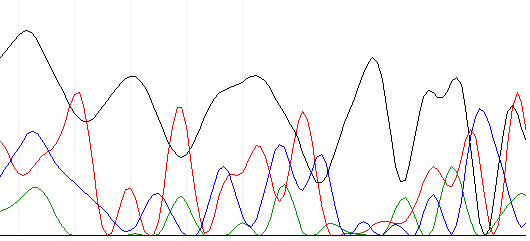
A couple days later, the sequencing results came back. The NAPS unit (the place we send our samples to be sequenced) said there was no signal from the results.
Below is an example of the chromatogram that came.
1,8-Cineole
Gurpal attempted to remove restriction sites from the PgxeTPS-Cin synthase gene, for Jacob, by doing a SDM-PCR. After doing it, he ran the PCR product through a gel and nothing showed up. Fortunately...
Jacob ran successful a SDM-PCR to remove a restriction site as verified by restriction digest and gel electrophoresis. He transformed competent cells with his SDM-PCR product and also ran SDM-PCR on another, closely related synthase (isolated from a hybrid between Picea glauca and Picea englmanii, as opposed to a pure Picea glauca). He ran Raf's mini-prep protocol on transformed cells to isolate a plasmid containing the SDMed pg-TPS-cin gene.
alpha-Pinene
New Pfu polymerases were used for Joe's SDM-PCR and it worked (using the Quikchange Site-directed Mutagensis kit )! Hurrah!
He found an optimal Quikchange PCR reaction mixture (for 50uL reaction):
| Reagents | Volume |
|---|---|
| ddH2O | 40uL |
| 10X Rxn Buffer | 5uL |
| dNTP (25mM | 1uL |
| Ps-TPS-Pin Fw(125ng) | 1uL |
| Ps-TPS-Pin Re(125ng) | 1uL |
| Pfu polymerase | 1uL |
| DNA template (50ng/uL) | 1uL |
Cycling Conditions:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95C | 1 min |
| 16 | 95C | 1 min |
| 55C | 1 min | |
| 68C | 10 min | |
| 1 | 4C | hold |
Before he moved on to the DpnI digest, Joe did a gel electrophoresis on the SDMed-PCR product. The band on the gel showed up as predicted (a pet200 plasmid at around 8.6kb). Joe then did a DpnI digest SDM-PCR products with 0.5uL of DpnI and incubated at 37C overnight. Afterwards, he transformed 2uL of DpnI products on Kanamycin plates. He also did a negative control to test the plates for kanamycin. Unfortunately, there is no growth of colonies in all plates. His next step is to increase the amount of DpnI digested products in the transformation.
At first, Joe didn't bother to do a gel electrophoresis on his PCR products after DpnI digest. He later did and found all his PCR products digest for some strange reason, which makes him wonder if he only had his original DNA template. His next step is to redo his SDM-PCR. So forget about the horray at the beginnning!
beta-Pinene & (-)-Limonene
Marianne and Vicki ran SDM-PCR on their synthase genes and verified their PCR products through gel electrophoresis. No bands were present in any of the lanes.
- Troubleshooting: The dNTP could have been too old/contaminated (we had trouble with this particular tube of dNTP from last year), the concentrations for the reagents were not optimized so... Marianne looked into some protocols and optimized the SDM-PCR protocol. Let the failures begin.
In a group, Vicki, Marianne, Gurpal and Jacob again attempted to perform SDM to remove an internal cut-site. ThermoPol was used as a buffer and PFU was used. After adding the primers and other constituents, we each ended up with ~ 25uL samples. We set our samples to run in a thermocycler for about 4 hours with specified conditions. When the cycling was completed, DpnI enzyme was added to each PCR tube and incubated overnight at 37oC.
To check if our SDM worked, we performed gel electrophoresis. Unfortunately, our gel showed that our site directed mutagenesis did not work as our product was still cut in two by the restriction enzyme. But, each of us learnt what not to do for next time which is always good.
 "
"