Team:British Columbia/Notebook/Week 14
From 2011.igem.org
| (19 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Notebook}} | {{Template:Notebook}} | ||
| + | <html> | ||
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b>Week 14: Sep 4-10</b><html><a name="w14"></a></html> | ||
===Poster Making=== | ===Poster Making=== | ||
| Line 11: | Line 15: | ||
| + | |||
| + | |||
| + | ===alpha-Pinene=== | ||
| + | |||
| + | Biobrick plasmids were sent to UBC NAPS sequencing. Results came back as a great sequence with Joe's illegal EcoRI site removed | ||
===Limonene Synthase=== | ===Limonene Synthase=== | ||
| Line 17: | Line 26: | ||
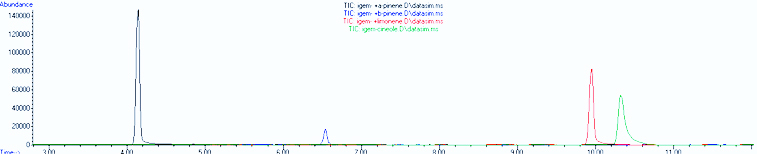
Joe and Alina went through some GC-MS data with Lina. We found out that the limonene synthase from the registry works! Read more about it on our <html><a href="https://2011.igem.org/Team:British_Columbia/Data">data page</a></html>. | Joe and Alina went through some GC-MS data with Lina. We found out that the limonene synthase from the registry works! Read more about it on our <html><a href="https://2011.igem.org/Team:British_Columbia/Data">data page</a></html>. | ||
| + | |||
| + | |||
| + | ===Smiley Face C41 Competent Cells=== | ||
| + | |||
| + | [[File:ubcigemsmiliecompetent.tif | left ]] | ||
| + | |||
| + | Jacob has made more C41 competent cells for characterization of the synthases in bacteria. | ||
| + | |||
| + | ===Cineole Synthase=== | ||
| + | Disaster! Jacob returned from a couple of weeks vacation back in his home town in Manitoba to find that sequencing showed that his biobrick construction didn't work! A PSTI site remained in the original PGXE synthase, and when the restriction digest and ligation were done, it cut out the part after the site. The sequencing results from the other synthase gene (the PG) showed a tyrosine kinase, which, while interesting, was not what we wanted to send in. Jacob went back to using site directed mutagenesis to remove that site from his original plasmid, having learned that you want to be sure of the sequence of a part before putting it in biobricks. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 23:17, 16 October 2011

 |
 |
 |
 |
 |
Contents |
Poster Making
Joe, Daisy and Marianne got together one evening to work on the poster. We realized after we finished making the template for the poster that we have no data! Alina also came into give her input on the poster.Later that night, Joe, Daisy, Marianne and Sam all went to Alina's lab to help her mash and pipet strawberry extract at 9 pm. A couple of Alina's lab members were also in the lab at 9 pm and were very surprised to find an illegal team of undergrads working on Alina's bench. PS We learned that Sam had a mullet and sang acapella at UCSF.
alpha-Pinene
Biobrick plasmids were sent to UBC NAPS sequencing. Results came back as a great sequence with Joe's illegal EcoRI site removed
Limonene Synthase
Joe and Alina went through some GC-MS data with Lina. We found out that the limonene synthase from the registry works! Read more about it on our data page.
Smiley Face C41 Competent Cells
Jacob has made more C41 competent cells for characterization of the synthases in bacteria.
Cineole Synthase
Disaster! Jacob returned from a couple of weeks vacation back in his home town in Manitoba to find that sequencing showed that his biobrick construction didn't work! A PSTI site remained in the original PGXE synthase, and when the restriction digest and ligation were done, it cut out the part after the site. The sequencing results from the other synthase gene (the PG) showed a tyrosine kinase, which, while interesting, was not what we wanted to send in. Jacob went back to using site directed mutagenesis to remove that site from his original plasmid, having learned that you want to be sure of the sequence of a part before putting it in biobricks.
 "
"