Team:British Columbia/Model3
From 2011.igem.org
| Line 24: | Line 24: | ||
<html><h3>Reactive site of the alpha-pinene synthase</h3></html><p></p> | <html><h3>Reactive site of the alpha-pinene synthase</h3></html><p></p> | ||
[[File:UBCiGEM skin dock.jpg]] | [[File:UBCiGEM skin dock.jpg]] | ||
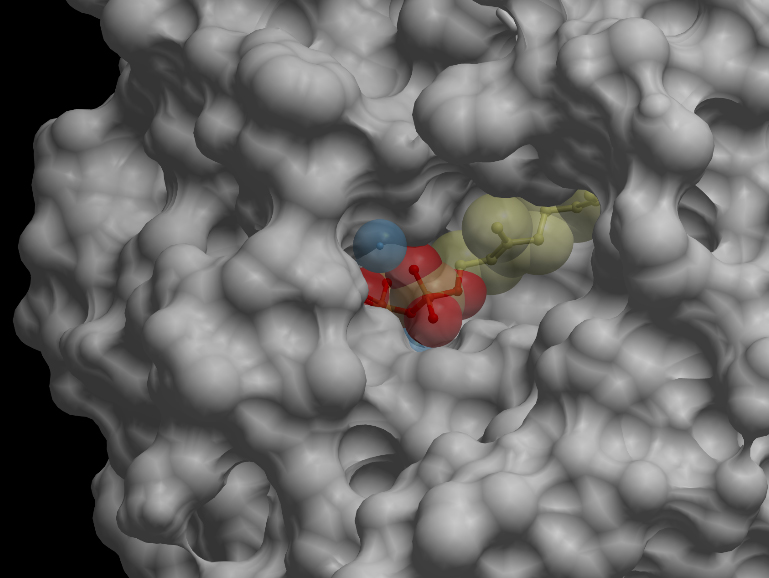
| - | Skin representation of the reactive site of alpha-pinene synthase. The substrate, geranyl diphosphate (GDP), is docked inside the reactive pocket. The oxygen atoms (red spheres) belonging to the phosphate groups of GDP and the carbon atoms (yellow spheres) forming the backbone of GDP are shown. Magnesium ion cofactors (blue spheres) interact with GDP.<p></p> | + | <p></p><b>Skin representation of the reactive site of alpha-pinene synthase. The substrate, geranyl diphosphate (GDP), is docked inside the reactive pocket. The oxygen atoms (red spheres) belonging to the phosphate groups of GDP and the carbon atoms (yellow spheres) forming the backbone of GDP are shown. Magnesium ion cofactors (blue spheres) interact with GDP.</b><p></p> |
[[File:UBCiGEM skeletal dock.jpg]] | [[File:UBCiGEM skeletal dock.jpg]] | ||
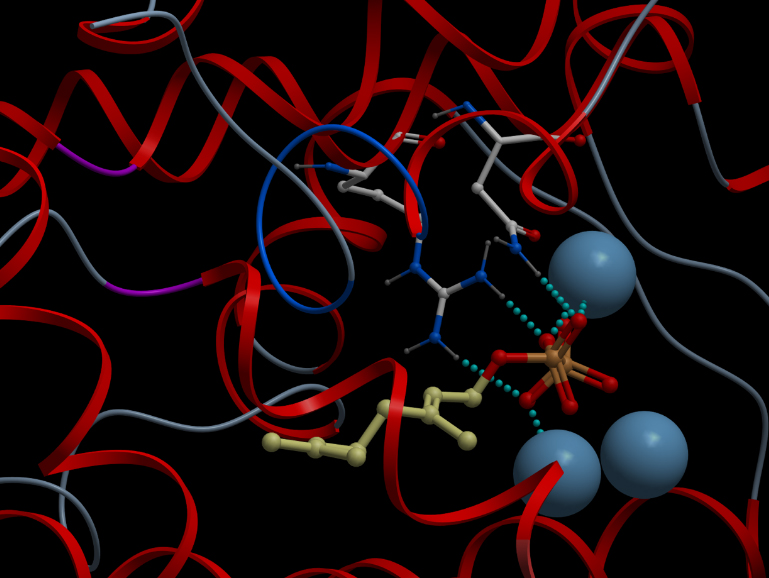
| - | Alternative representation of the reactive site of alpha-pinene synthase. The synthase structure is shown in ribbon model and the substrate GDP in ball-and-stick model. The magnesium ion cofactors are represented by blue spheres. The dotted light blue lines indicate hydrogen bonds.<p></p> | + | <p></p><b>Alternative representation of the reactive site of alpha-pinene synthase. The synthase structure is shown in ribbon model and the substrate GDP in ball-and-stick model. The magnesium ion cofactors are represented by blue spheres. The dotted light blue lines indicate hydrogen bonds.</b><p></p> |
<html><h3>Materials and Methods</h3></html><p></p> | <html><h3>Materials and Methods</h3></html><p></p> | ||
| - | + | We used MODELLER (1) to automate homology-based 3D structure prediction. We identified an experimentally determined 3D structure of a taxadiene synthase from Pacific Yew (PDB ID: 3P5R) as an appropriate template. All images were taken using the ICM by Molsoft LLC (2). | |
<html><h3>Future Directions</h3></html><p></p> | <html><h3>Future Directions</h3></html><p></p> | ||
| - | + | The current study a | |
<html><h3>References</h3></html><p></p> | <html><h3>References</h3></html><p></p> | ||
1. N. Eswar, M. A. Marti-Renom, B. Webb, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper, A. Sali. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1-5.6.30, 2006. | 1. N. Eswar, M. A. Marti-Renom, B. Webb, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper, A. Sali. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1-5.6.30, 2006. | ||
| + | 2. Abagyan, R.A., Totrov, M.M., and Kuznetsov, D.A. Icm: A New Method For Protein Modeling and Design: Applications To Docking and Structure Prediction From The Distorted Native Conformation. J. Comp. Chem. 15, 488-506. 1994. | ||
| + | |||
| + | <html><h3>Acknowledgment</h3></html><p></p> | ||
| + | We are very grateful to Yvonne Y. Li from the BC Genome Sciences Centre for valuable technical advice. | ||
Revision as of 03:50, 29 October 2011

Monoterpene Synthase Structural Modeling
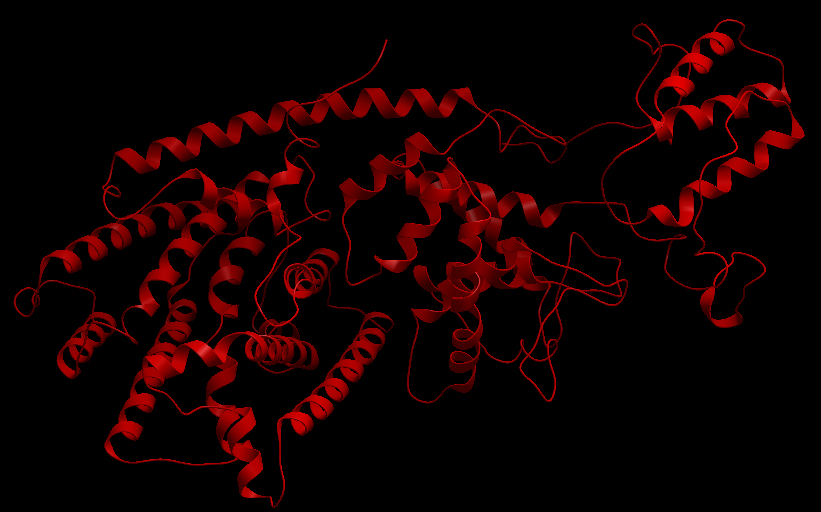
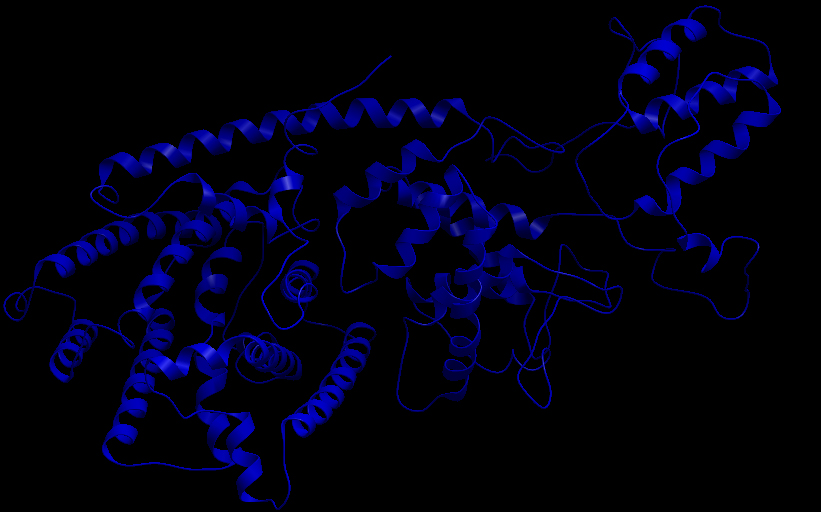
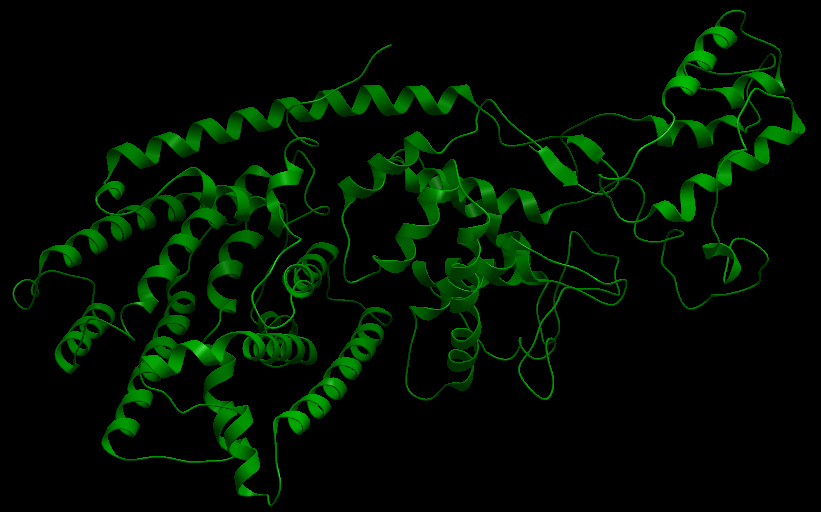
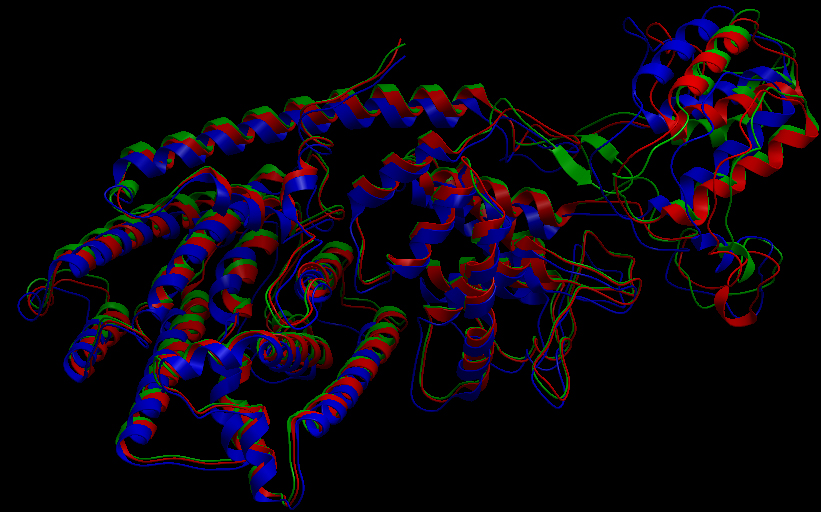
Three-dimensional Structure of the 3 Terpene Synthase Proteins
Ribbon representation of the alpha-pinene synthase. Ribbon representation of the beta-pinene synthase. Ribbon representation of the limonene synthase. Superimposition of the 3D structures of the three synthase (blue), beta-pinene synthase (red), and limonene synthase (green). The synthases exhibit high structural similarity. This observation allows us to identify homologous amino acid residues that may affect terpene synthesis efficiency.Reactive site of the alpha-pinene synthase
Skin representation of the reactive site of alpha-pinene synthase. The substrate, geranyl diphosphate (GDP), is docked inside the reactive pocket. The oxygen atoms (red spheres) belonging to the phosphate groups of GDP and the carbon atoms (yellow spheres) forming the backbone of GDP are shown. Magnesium ion cofactors (blue spheres) interact with GDP. Alternative representation of the reactive site of alpha-pinene synthase. The synthase structure is shown in ribbon model and the substrate GDP in ball-and-stick model. The magnesium ion cofactors are represented by blue spheres. The dotted light blue lines indicate hydrogen bonds.Materials and Methods
We used MODELLER (1) to automate homology-based 3D structure prediction. We identified an experimentally determined 3D structure of a taxadiene synthase from Pacific Yew (PDB ID: 3P5R) as an appropriate template. All images were taken using the ICM by Molsoft LLC (2).
Future Directions
The current study a
References
1. N. Eswar, M. A. Marti-Renom, B. Webb, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper, A. Sali. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1-5.6.30, 2006. 2. Abagyan, R.A., Totrov, M.M., and Kuznetsov, D.A. Icm: A New Method For Protein Modeling and Design: Applications To Docking and Structure Prediction From The Distorted Native Conformation. J. Comp. Chem. 15, 488-506. 1994.
Acknowledgment
We are very grateful to Yvonne Y. Li from the BC Genome Sciences Centre for valuable technical advice.
 "
"