Team:British Columbia/Model3
From 2011.igem.org
(Difference between revisions)
| Line 11: | Line 11: | ||
<html><h3>Three-dimensional structure of the 3 terpene synthases</h3></html><p></p> | <html><h3>Three-dimensional structure of the 3 terpene synthases</h3></html><p></p> | ||
[[File:UBCiGEM superimposed synthases.jpg]] | [[File:UBCiGEM superimposed synthases.jpg]] | ||
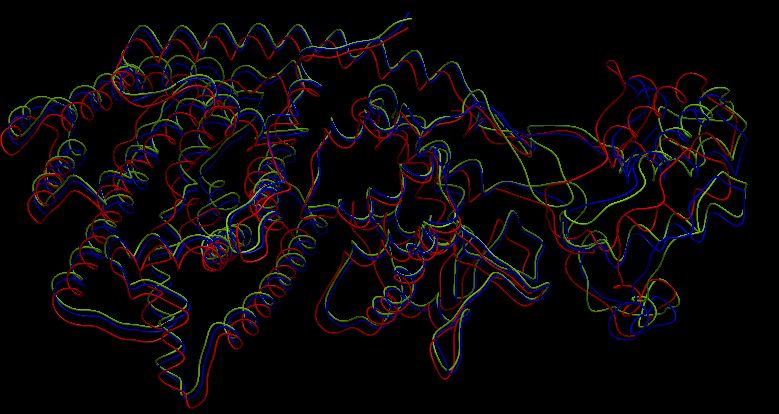
| - | <p></p>Superimposition of the 3D structures of alpha-pinene synthase (blue), beta-pinene synthase (red), and limonene synthase (green). We used MODELLER (1) to automate homology-based 3D structure prediction. We identified an experimentally determined 3D structure of a taxadiene synthase from Pacific Yew (PDB ID: 3P5R) as an appropriate template.<p></p> | + | <p></p>Superimposition of the 3D structures of alpha-pinene synthase (blue), beta-pinene synthase (red), and limonene synthase (green). We used MODELLER (1) to automate homology-based 3D structure prediction. We identified an experimentally determined 3D structure of a taxadiene synthase from Pacific Yew (PDB ID: 3P5R) as an appropriate template. The image was taken using the freely available ICM Browser.<p></p> |
<b>Reactive site of the three synthases</b><p></p> | <b>Reactive site of the three synthases</b><p></p> | ||
Revision as of 02:41, 29 October 2011

Monoterpene Synthase Structural Modeling
Three-dimensional structure of the 3 terpene synthases
Superimposition of the 3D structures of alpha-pinene synthase (blue), beta-pinene synthase (red), and limonene synthase (green). We used MODELLER (1) to automate homology-based 3D structure prediction. We identified an experimentally determined 3D structure of a taxadiene synthase from Pacific Yew (PDB ID: 3P5R) as an appropriate template. The image was taken using the freely available ICM Browser. Reactive site of the three synthases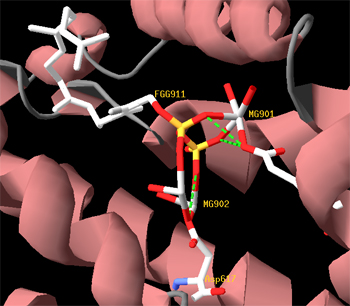 Magnesium ions are cofactors located at the reactive site.
Magnesium ions are cofactors located at the reactive site.
Future Directions
References
1. N. Eswar, M. A. Marti-Renom, B. Webb, M. S. Madhusudhan, D. Eramian, M. Shen, U. Pieper, A. Sali. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement 15, 5.6.1-5.6.30, 2006.
 "
"