Team:British Columbia/Notebook/Week 8
From 2011.igem.org
Watermelon (Talk | contribs) |
|||
| Line 45: | Line 45: | ||
Joe finally received his primers for PCR amplification of his SDM-alpha pinene synthase necessary for the ligation into yeast and biobrick plasmids. He did a PCR using these primers. However, no PCR products were seen after gel verification. The annealing temperature may have been too low at 55 degrees Celsius. Therefore, he needs to increase the annealing temperature to 60-72 degrees Celsius, the optimal temperature for Phusion Hot Start Polymerase. | Joe finally received his primers for PCR amplification of his SDM-alpha pinene synthase necessary for the ligation into yeast and biobrick plasmids. He did a PCR using these primers. However, no PCR products were seen after gel verification. The annealing temperature may have been too low at 55 degrees Celsius. Therefore, he needs to increase the annealing temperature to 60-72 degrees Celsius, the optimal temperature for Phusion Hot Start Polymerase. | ||
| + | |||
| + | '''IDI1, HMG2 metabolic genes''' | ||
| + | |||
| + | Sam performed PCR on one sample of HMG2 plasmid and two samples of IDI1 plasmid to add an SMA1 cutsite upstream of the promoter on both genes. This PCR was unsuccessful; no bands were seen for any of the unpurified PCR product. | ||
Revision as of 22:36, 6 August 2011

 |
 |
 |
 |
 |
3-Carene
Daisy has been PCRing out the synthase with the yeast primers that contain the NotI and SmaI cut sites and an N-terminal His-tag. However, the forward primer is quite long and Daisy thinks there is some nonspecific annealing happening.
ERG20
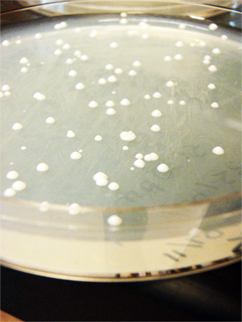
Gurpal performed a yeast transformation. He separately transformed pBS and pKS into wildtype yeast. It was a success!
(-)-limonene Results from transformation:
- Positive control: TMTC
- Negative control: no colonies
- pADM743: TMTC
Set up O/N culture for mini-prepping
- 4 tubes: 3 colonies picked + 1 control
Beta-pinene
Marianne's transformations (yeast plasmids + synthase) show a few colonies each on 3 plates (original-his-GAL, SDM-no-his-GPD, SDM-no-his-GAL. Marianne conducted a colony PCR of these yeast plasmid and synthase transformants and gel verified them. The gel image shows bands at the correct lengths (~500bp), indicating that the ligations were successful! We have beta-pinene on yeast plasmids!
Marianne started on the Biobrick assembly; she restriction digested and ligated synthase and pSB1C3 backbone. Marianne set up an O/N culture for the yeast plasmids. Marianne transformed the Biobrick ligations and miniprepped the yeast plasmids. Marianne also transformed the miniprepped yeast plasmids(SND, SNL) into wildtype yeast, pKS yeast, pBS yeast.
1,8-cineole
The previous colonies of E. coli that should have had the 1,8-cineole gene on pSB1C3 were shown to have different sized plasmids after overnight cultures and minipreps. Of the 5 colonies, 4 had plasmids a little over 2 kb in size, and one had a plasmid around 5 kb in size. The 1,8-cineole gene is 1.7 kb and the pSB1C3 plasmid that is left after the restriction digest is 2 kb, so the product should have been around 3.7 kb. A restriction digest with ECORI and PSTI showed only one band for the 5 kb product, and the 2 kb product showed bands at about 1.7 kb and 700 bp. All of these were grown on chlor plates and chlor LB broth for the overnight culture. A gel of just the pSB1C3 showed a product well below the 500 bp mark on 1 kb ladder.
This was rather disconcerting, so Jacob decided to run another biobrick PCR and try the ligation again.
Alpha-pinene synthase
Joe finally received his primers for PCR amplification of his SDM-alpha pinene synthase necessary for the ligation into yeast and biobrick plasmids. He did a PCR using these primers. However, no PCR products were seen after gel verification. The annealing temperature may have been too low at 55 degrees Celsius. Therefore, he needs to increase the annealing temperature to 60-72 degrees Celsius, the optimal temperature for Phusion Hot Start Polymerase.
IDI1, HMG2 metabolic genes
Sam performed PCR on one sample of HMG2 plasmid and two samples of IDI1 plasmid to add an SMA1 cutsite upstream of the promoter on both genes. This PCR was unsuccessful; no bands were seen for any of the unpurified PCR product.
 "
"