Team:British Columbia/Accomplishments
From 2011.igem.org
| (159 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Template_HD_1}} | {{Template:Template_HD_1}} | ||
| + | <html> | ||
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"></html> | ||
| - | < | + | <font size="+3"><font color="green">Wet Laboratory Achievements</font></font> |
| - | + | <hr style="color:#BDCBBD; height:3px;" /> | |
| - | + | ||
| - | </ | + | |
| - | + | ||
| - | + | ||
| - | < | + | <b></b> |
| - | + | <font size="2"><h3>In vitro assay production of monoterpene in bacteria</h3></font> | |
| - | + | ||
| - | + | <b></b><font size="1"><h3>Geranyl Pyrophosphate (GPP) Assay</h3></font> | |
| - | < | + | |
| - | < | + | Alpha-pinene and Beta-pinene synthases were purified using His SpinTrap Ni-affinity columns and were assayed in vitro with GPP. Using gas chromatography-mass spectrometry (GCMS), we confirmed the synthesis of alpha and beta pinene monoterpenes from our enzyme assays. |
| - | < | + | |
| - | </ | + | <b></b><font size="1"><h3>GC-MS Chromatogram</h3></font> |
| - | + | <center>[[File:UBCiGEM_GC_beta_pinene.jpg]]</center> | |
| - | + | ||
| - | </br>< | + | |
| - | + | <b></b> | |
| - | + | <font size="2"><h3>Diterpene production in yeast</h3></font> | |
| - | </ | + | |
| - | + | Levoabietadiene synthase was expressed in yeast and an in vivo assay was done. The samples were analyzed with GCMS. The products, abietadiene, neoabietadiene, levopimaradiene were produced. This shows that terpenes can be produced in yeast. | |
| - | + | ||
| - | a | + | |
| - | + | <b></b><font size="1"><h3>GC-MS Chromatogram</h3></font> | |
| - | </br></br> | + | <center>[[File:UBCiGEM_GC_abidiene.jpg]]</center> |
| - | + | <b></b> | |
| - | + | <font size="2"><h3>Mountain Pine Beetle & Yeast Co-culture</h3></font> | |
| - | </br></ | + | |
| - | <h3> | + | To prove the effectiveness of our theoretical release model of Saccharomyces cerevisiae into the wild, we investigated whether wild-type yeast will survive during the transporation process (via the mountain pine beetles) from plate of origin (plate with yeast products; simulating our trapbox) to the next media plate. Beetles were incubated with yeast and were challenged with diferent amount of times away from the next media plates (in empty plates for 0, 10, 24, and 36 hours of time; simulating the transfer period when beetles leave the trapbox and fly off to the next tree). Results were analyzed qualitatively for the presence of GFP after growth on selective media plates from each time challenge. Based on these results, we can conclude that pine beetles do can carry wild-type yeast and transfer them onto the next media plate. It appears that our yeast product can survive on the beetles for 36 hours away from media plate. However, it cannot be determined whether the amount of yeast products on the beetles declines with time. |
| + | |||
| + | |||
| + | <html><center><iframe width="425" height="349" src="http://www.youtube.com/embed/-fXdy8plAR8?hl=en&fs=1" frameborder="0" allowfullscreen></iframe></center></html> | ||
| + | |||
| + | <center>[[File:Yst-MPB.jpg]]</center> | ||
| + | <b></b> | ||
| + | <font size="2"><h3>Blue Stain Fungi & Yeast Co-culture</h3></font> | ||
| + | |||
| + | To determine whether terpene producing S.cerevisiae can grow with and/or inhibit the growth of G. clavigera, we co-cultured blue stain fungi with diterpene producing yeast and wild-type yeast in the same media. However, results are inclusive and we are currently optomizing the protocol for co-culturing. | ||
| + | |||
| + | <center>[[File:3cocultureubcigem2011.jpg]]</center> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <font size="+3"><font color="green">Modeling Achievements</font></font> | ||
| + | <hr style="color:#BDCBBD; height:3px;" /> | ||
| + | <b></b> | ||
| + | <font size="2"><h3>Monoterpene Production Model</h3></font> | ||
| + | |||
| + | |||
| + | We modeled a simplified and modified version of the mevalonate pathway that describes our engineered yeast cells. We created a series of differential equations to model each chemical reaction that were based on first-order Michaelis-Menten kinetics. Enzyme constants were estimated/found using literature values and the simulations were conducted using MATLAB. | ||
| + | |||
| + | Our simulation showed that 17.10% more beta-pinene is produced and 17.00 % more alpha-pinene is produced when K6R-hmg2 and erg20-2 are used instead of HMG2 and ERG20 alone in a yeast cell. For manufacturing purposes, sensitivity analysis was performed and it was determined that the pathway could be improved to increase the production of monoterpenes by increasing the concentration of the enzymes K6R-HMG2 and IDI1 for DMA-PP. In particular, the tripling the [K6R-HMG2] increases the [GPP] by 8.7000 times. The tripling the [K6R-HMG2] and [IDI1] for the production of DMA-PP increases the [B-pinene] by 7.3166 times and 1.3052 times respectively. | ||
| + | |||
| + | <center>[[File:Monoterpene Production vs Time in Pinene Synthase.jpg]]</center> | ||
| + | |||
| + | <b></b> | ||
| + | <font size="2"><h3>Beetle Epidemic Model</h3></font> | ||
| + | |||
| + | We employed a cluster-based modeling to predict the spread of mountain pine beetle infestation from year 2011 up to 2020 based on cluster analysis. In addition, we identified strategic subpopulation centres for deployment of synthetic products. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <center><iframe width="480" height="360" src="http://www.youtube.com/embed/SsZUnF6Y6yw" frameborder="0" allowfullscreen></iframe></center> | ||
| + | </br></br></div> | ||
| + | <div id="links"> | ||
| + | |||
| + | <b></b> | ||
| + | <font size="2"><h3>Synthase Model</h3></font> | ||
| + | |||
| + | We constructed 3D structure models of alpha-pinene, beta-pinene, and limonene synthase to identify amino-acid residues that can be modified in the future to improve terpene productions. | ||
| + | |||
| + | <center>[[File:UBCiGEM skeletal dock.jpg]]</center> | ||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="+3"><font color="green">Human Practices Achievements</font></font> | ||
| + | <hr style="color:#BDCBBD; height:3px;" /> | ||
| + | |||
| + | <b></b> | ||
| + | <font size="2"><h3>High School Mentorship</h3></font> | ||
| + | |||
| + | We collaborated with Science World and Future Leaders in Science to deliver a synthetic biology workshop for local high school students. During our workshop, we introduced the idea of synthetic biology and iGEM to the high school students and guided them through a <html><a href="http://openwetware.org/wiki/IGEM_Outreach:Plasmid_Activity">plasmid activity</a></html> aimed at solving a real world problem by creating a plasmid with appropriate parts. Our workshop outline can be found at the wiki for <html><a href="http://openwetware.org/wiki/IGEM_Outreach:Life_in_the_Lab">iGEM outreach</a></html>. We also created a guide for building a high school iGEM team, which can be found <html><a href="https://static.igem.org/mediawiki/2011/a/ad/TeamBritishColumbiasGuidetoStartaHigh-schooliGEMteam.pdf">downloaded</a></html> or found on <html><b><a href="http://openwetware.org/wiki/IGEM_Outreach:Life_in_the_Lab">the CommunityBricks page</a></b></html>. | ||
| + | |||
| + | <html></br><center><img src="https://static.igem.org/mediawiki/2011/b/b9/Ubcigemscienceworldoutreach.jpg" width=800px> | ||
| + | </center></html> | ||
| + | |||
| + | <b></b> | ||
| + | <font size="2"><h3>Synthetic Biology Gone Wild</h3></font> | ||
| + | |||
| + | Our project has the prospect of introducing synthetic organisms into the environment, which is a controversial idea. First of all, we initiated an open dialogue, "Synthetic Biology in the Open: Pipe dream or the next giant leap for mankind?" which can be <html><a href="https://static.igem.org/mediawiki/2011/4/45/Ubcigem2011syntheticbiologyintheopen.pdf">downloaded</a></html> or found on <html><a href="http://openwetware.org/wiki/IGEM_Outreach:Synthetic_Biology_in_the_Open">this CommunityBricks page</a>.''' | ||
| + | |||
| + | Next we conducted interviews with experts in our community to understand what the current attitudes are towards releasing synthetic biology in the wild. | ||
| - | |||
| - | < | + | <center><img src="https://static.igem.org/mediawiki/2011/f/fe/UBCiGEM_skeletal_dock.jpg" width="50%" height="50%"</center> |
| + | <b></b></center> | ||
| + | <font size="2"><h3>Public Perceptions of Synthetic Biology</h3></font></html> | ||
| + | From UBC orientation day, we collected students' perceptions on synthetic biology (presented in a word cloud)and observed whether it has changed for the past years. | ||
| - | |||
| - | <html>< | + | <html><h4>What is Synthetic Biology?</h4> |
| - | + | <iframe id="xtranormal_What is Synthetic Biology?" name="xtranormal_What is Synthetic Biology?" style="width:480px;height:299px;" src="http://www.xtranormal.com/xtraplayr/12434771/what-is-synthetic-biology" marginwidth="0" marginheight="0" border="0" frameborder="0" scrolling="auto"></iframe> | |
| - | + | <img src="https://static.igem.org/mediawiki/2011/3/39/Ubcigemwordcloud2010.jpg" width="25%" height="25%" align="right" hspace="100"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | </html> | |
Latest revision as of 03:59, 29 October 2011

Wet Laboratory Achievements
In vitro assay production of monoterpene in bacteria
Geranyl Pyrophosphate (GPP) Assay
Alpha-pinene and Beta-pinene synthases were purified using His SpinTrap Ni-affinity columns and were assayed in vitro with GPP. Using gas chromatography-mass spectrometry (GCMS), we confirmed the synthesis of alpha and beta pinene monoterpenes from our enzyme assays.
GC-MS Chromatogram
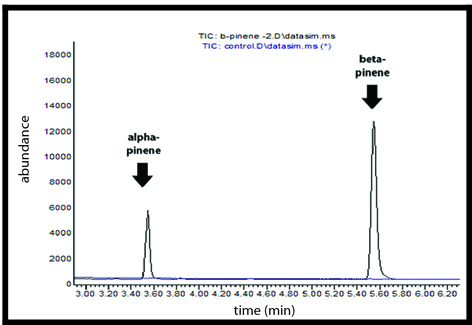
Diterpene production in yeast
Levoabietadiene synthase was expressed in yeast and an in vivo assay was done. The samples were analyzed with GCMS. The products, abietadiene, neoabietadiene, levopimaradiene were produced. This shows that terpenes can be produced in yeast.
GC-MS Chromatogram
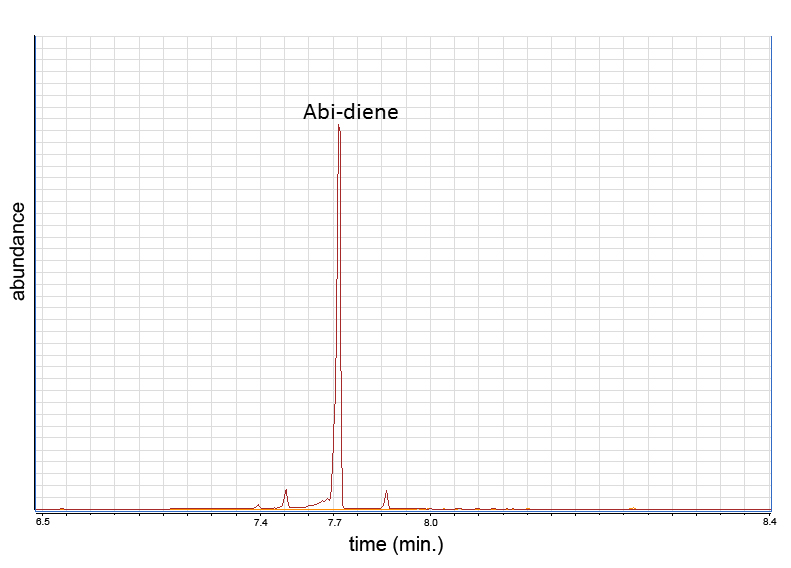
Mountain Pine Beetle & Yeast Co-culture
To prove the effectiveness of our theoretical release model of Saccharomyces cerevisiae into the wild, we investigated whether wild-type yeast will survive during the transporation process (via the mountain pine beetles) from plate of origin (plate with yeast products; simulating our trapbox) to the next media plate. Beetles were incubated with yeast and were challenged with diferent amount of times away from the next media plates (in empty plates for 0, 10, 24, and 36 hours of time; simulating the transfer period when beetles leave the trapbox and fly off to the next tree). Results were analyzed qualitatively for the presence of GFP after growth on selective media plates from each time challenge. Based on these results, we can conclude that pine beetles do can carry wild-type yeast and transfer them onto the next media plate. It appears that our yeast product can survive on the beetles for 36 hours away from media plate. However, it cannot be determined whether the amount of yeast products on the beetles declines with time.
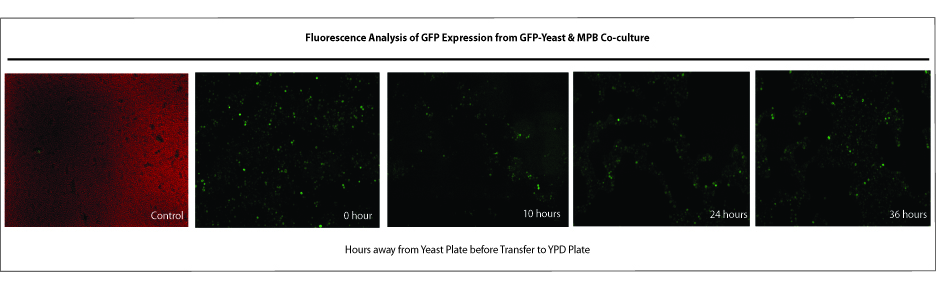
Blue Stain Fungi & Yeast Co-culture
To determine whether terpene producing S.cerevisiae can grow with and/or inhibit the growth of G. clavigera, we co-cultured blue stain fungi with diterpene producing yeast and wild-type yeast in the same media. However, results are inclusive and we are currently optomizing the protocol for co-culturing.
Modeling Achievements
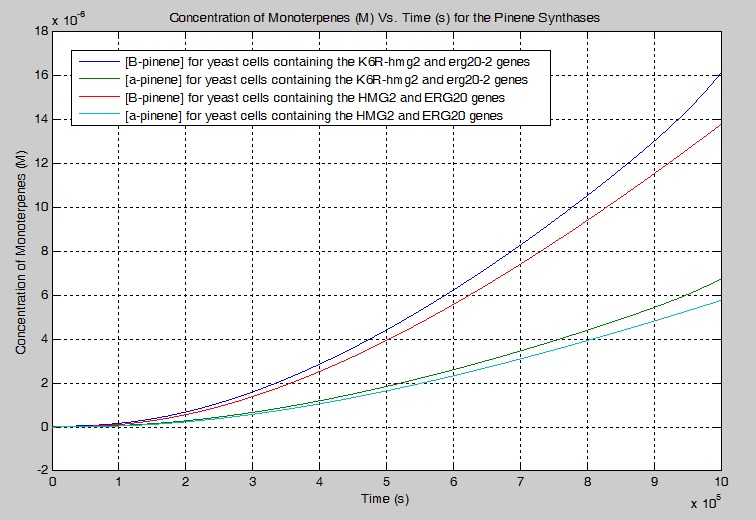
Monoterpene Production Model
We modeled a simplified and modified version of the mevalonate pathway that describes our engineered yeast cells. We created a series of differential equations to model each chemical reaction that were based on first-order Michaelis-Menten kinetics. Enzyme constants were estimated/found using literature values and the simulations were conducted using MATLAB.
Our simulation showed that 17.10% more beta-pinene is produced and 17.00 % more alpha-pinene is produced when K6R-hmg2 and erg20-2 are used instead of HMG2 and ERG20 alone in a yeast cell. For manufacturing purposes, sensitivity analysis was performed and it was determined that the pathway could be improved to increase the production of monoterpenes by increasing the concentration of the enzymes K6R-HMG2 and IDI1 for DMA-PP. In particular, the tripling the [K6R-HMG2] increases the [GPP] by 8.7000 times. The tripling the [K6R-HMG2] and [IDI1] for the production of DMA-PP increases the [B-pinene] by 7.3166 times and 1.3052 times respectively.
Beetle Epidemic Model
We employed a cluster-based modeling to predict the spread of mountain pine beetle infestation from year 2011 up to 2020 based on cluster analysis. In addition, we identified strategic subpopulation centres for deployment of synthetic products.
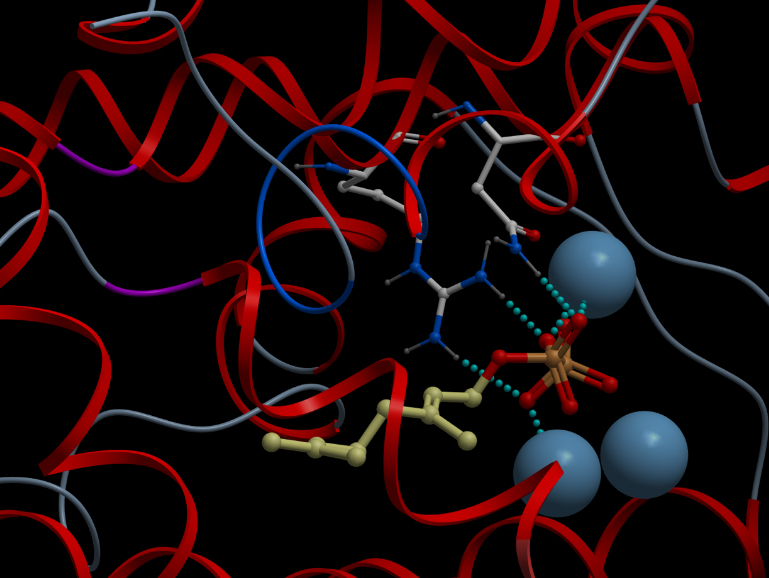
Synthase Model
We constructed 3D structure models of alpha-pinene, beta-pinene, and limonene synthase to identify amino-acid residues that can be modified in the future to improve terpene productions.Human Practices Achievements
High School Mentorship
We collaborated with Science World and Future Leaders in Science to deliver a synthetic biology workshop for local high school students. During our workshop, we introduced the idea of synthetic biology and iGEM to the high school students and guided them through a plasmid activity aimed at solving a real world problem by creating a plasmid with appropriate parts. Our workshop outline can be found at the wiki for iGEM outreach. We also created a guide for building a high school iGEM team, which can be found downloaded or found on the CommunityBricks page.
Synthetic Biology Gone Wild
Our project has the prospect of introducing synthetic organisms into the environment, which is a controversial idea. First of all, we initiated an open dialogue, "Synthetic Biology in the Open: Pipe dream or the next giant leap for mankind?" which can be downloaded or found on this CommunityBricks page.''' Next we conducted interviews with experts in our community to understand what the current attitudes are towards releasing synthetic biology in the wild.
Public Perceptions of Synthetic Biology
From UBC orientation day, we collected students' perceptions on synthetic biology (presented in a word cloud)and observed whether it has changed for the past years.
What is Synthetic Biology?

 "
"