Team:British Columbia/Model1
From 2011.igem.org
(→MODEL 1: Secretion and Production of Monoterpenes in Yeast) |
|||
| Line 36: | Line 36: | ||
These are the differential equations that are common to each biochemical pathway: | These are the differential equations that are common to each biochemical pathway: | ||
| - | [[File: | + | [[File:Common_DE.jpg | frame | center | Differential equations common in all products. ]] |
Since each synthase gene combines with GPP to produce different monoterpenes, some differential equations will be specific to a certain synthase. | Since each synthase gene combines with GPP to produce different monoterpenes, some differential equations will be specific to a certain synthase. | ||
Revision as of 18:16, 22 September 2011

MODEL 1: Secretion and Production of Monoterpenes in Yeast
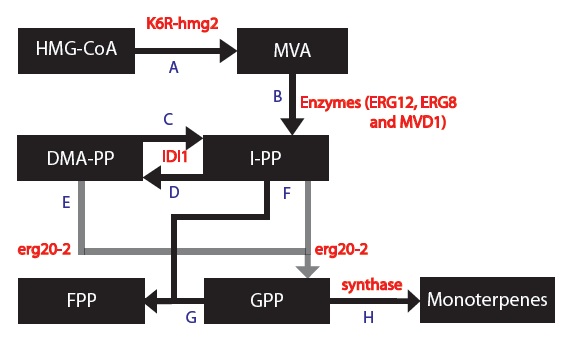
OBJECTIVE: To create a model to assay the production of monoterpenes in our genetically engineered yeast cells given that they contain the metabolic genes IDI1, K6R-hmg2, erg20-2 and a synthase gene (one of PgTPS-Pin2, PsTPS-Car1, PgxeTPS-Cin, PgTPS-Cin and PsTPS-Pin).
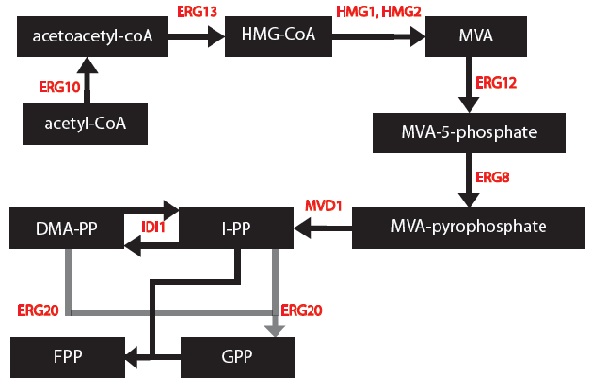
My goal was to create a model to assay the production of monoterpenes in our genetically engineered yeast cells given that they contain the metabolic genes IDI1, K6R-hmg2, erg20-2 and a synthase gene (one of PgTPS-Pin2, PsTPS-Car1, PgxeTPS-Cin, PgTPS-Cin and PsTPS-Pin). First, a series of block diagrams to model the biochemical pathways were created. To illustrate, I started with the mevalonate pathway (Ignea, et al., 2011) and simplified the pathway to only relevant enzymes associated with our project – the IDI1, HMG2 and ERG20. Since each yeast cell contained a different synthase gene, the end of each biochemical pathway was modified accordingly to produce to correct monoterpenes (Keeling, et al., 2011). Next, a series of chemical equations were created. For instance, 6 chemical reactions are common to each yeast cells. This part was important because I began to note the kinetic constants. Afterwards, I created differential equations based on the simplified biochemical pathways and chemical reactions. Dr. Eric Lagally referred me to 2 sources to help me make this conversion: “Writing Differential Equations” (Bourne, 2010) and “Chapter 6- Transport and Kinetics” (Jakubowski, 2011). The differential equations I made were based on using first order and Michelson-menten kinetics. At this point, many constants and concentrations must be known to solve the differential equations. In particular, there is an enzyme database called “BRENDA” which contains some of the values – kcat and km (Scheer, 2011). Our project is based on research that has been conducted in 2011 so some variables will need to be experimentally determined. The solutions to these differential equations will be solved in MATLAB and plots of concentration of monterpene as a function of substrate or time will be created.
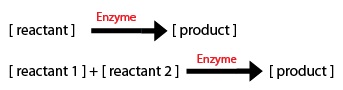
We made a list of important common chemical reactions after developing this simplified version of the biochemical pathway:
Since each synthase gene combines with GPP to produce different monoterpenes, some chemical reactions will be specific to a certain synthase.
Most of the equations fall under the main 2 types of Michaelis–Menten equations:
Next, we created differential equations from the series of chemical reactions and block diagrams for each of the biochemical pathways.
These are the differential equations that are common to each biochemical pathway:
Since each synthase gene combines with GPP to produce different monoterpenes, some differential equations will be specific to a certain synthase.
The differential equations specific to the PgTPS-Pin2 synthase gene:
The differential equations specific to the PsTPS-Car1 synthase gene:
The differential equations specific to the PgxeTPS-Cin synthase gene:
The differential equations specific to the PgTPS-Cin synthase gene:
The differential equations specific to the PsTPS-Pin synthase gene:
 "
"