Team:British Columbia/Notebook/Week 8
From 2011.igem.org
(Created page with "On July 25th, Gurpal performed a yeast transformation. He transformed his purified pBS and pKS into 2 separate wildtype yeasts. He followed a protocol, left the cells to incubate...") |
|||
| (55 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Template:Notebook}} | |
| + | <html> | ||
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b> Week 8: July 24-30</b> <html><a name="w8"></a></html> | ||
| - | + | ==Lab Meeting - July 25== | |
| + | Not a lot happened at today's meeting. | ||
| + | |||
| + | Due to safety reasons, a reminder that everyone needs to be in pairs, especially after the typical working hours (9am-5pm). | ||
| + | |||
| + | [[File:Minmeetwk8ubc.JPG | thumb | 300px]] | ||
| + | |||
| + | ==Presentation== | ||
| + | |||
| + | The team got together to discuss the presentation. We talked about what we liked and didn't like about the presentations we watched. For example, we didn't want all team members to be on the platform with the presenters because it feels distracting and unnecessary. Also, some of us thought that the flow of the story is very important; powerpoint slides shouldn't be consumed by wordy sentences but with clear and simple pictures with few words. | ||
| + | In the end, we agreed that there would be 2-3 presenters. | ||
| + | |||
| + | We discussed the criteria to evaluate the potential presenters; the plan is for potential presenters to present a powerpoint on their role in the project, and feedback will be given at the end by everyone. | ||
| + | |||
| + | ==Constructing the Wiki== | ||
| + | Vicki, Alina, and Gurpal started thinking of template ideas for the wiki. Because Vicki cannot draw or use photoshop to save her life, Alina and Gurpal volunteered to create most (if not all) of the images for the wiki. You know what this means: drawing contest... although Gurpal calls it, "Alina Wins!" | ||
| + | |||
| + | In the meantime, Vicki downloaded the trial version of Dreamweaver and began looking through the online tutorials. Her goal is to master Dreamweaver so that she can use it to design the format of the wiki. Let the late nights begin. | ||
| + | |||
| + | ==3-Carene== | ||
| + | |||
| + | Daisy has been PCRing out the synthase with the yeast primers that contain the NotI and SmaI cut sites and an N-terminal His-tag. However, the forward primer is quite long and Daisy thinks there is some nonspecific annealing happening. | ||
| + | |||
| + | ==1,8-Cineole== | ||
| + | |||
| + | The previous colonies of <i>E. coli</i> that should have had the 1,8-cineole synthase gene on pSB1C3 were shown to have different sized plasmids after overnight cultures and minipreps. Of the 5 colonies, 4 had plasmids a little over 2 kb in size, and one had a plasmid around 5 kb in size. The 1,8-cineole gene is 1.7 kb and the pSB1C3 plasmid that is left after the restriction digest is 2 kb, so the product should have been around 3.7 kb, suggestion it didn't work. A restriction digest with ECORI and PSTI showed only one band for the 5 kb product, and the 2 kb product showed bands at about 1.7 kb and 700 bp. | ||
| + | |||
| + | These results made no sense, so to check if there was a problem with the plasmid, pure pSB1C3 was run on an agarose gel. The gel showed a product well below the 500 bp mark on a 1 kb ladder. | ||
| + | |||
| + | This was rather disconcerting, so Jacob decided to run another biobrick PCR and try the ligation again with another source of the plasmid. | ||
| + | |||
| + | ==alpha-Pinene== | ||
| + | |||
| + | Joe finally received his primers for PCR amplification of his SDM-alpha pinene synthase necessary for the ligation into yeast and biobrick plasmids. He did a PCR using these primers. However, no PCR products were seen after gel verification. The annealing temperature may have been too low at 55 degrees Celsius. Therefore, he needs to increase the annealing temperature to 60-72 degrees Celsius, the optimal temperature for Phusion Hot Start Polymerase. | ||
| + | |||
| + | ==beta-Pinene== | ||
| + | |||
| + | [[File:mariyeast.jpg | thumb | left | 200px | Marianne's transformants]] | ||
| + | |||
| + | Marianne's transformations (yeast plasmids + synthase) show a few colonies each on 3 plates (original-his-GAL, SDM-no-his-GPD, SDM-no-his-GAL. Marianne conducted a colony PCR of these yeast plasmid and synthase transformants and gel verified them. The gel image shows bands at the correct lengths (~500bp), indicating that the ligations were successful! We have beta-pinene on yeast plasmids! | ||
| + | |||
| + | Marianne started on the Biobrick assembly; she restriction digested and ligated synthase and pSB1C3 backbone. Marianne set up an O/N culture for the yeast plasmids. Marianne transformed the Biobrick ligations and miniprepped the yeast plasmids. Marianne also transformed the miniprepped yeast plasmids(SND, SNL) into wildtype yeast, pKS yeast, pBS yeast. | ||
| + | |||
| + | <br><br> | ||
| + | ==(-)-Limonene== | ||
| + | |||
| + | [[File:Gurpalyeast1.jpg | thumb | right | 150px | Gurpal's yeast cultures]] [[File:Gurpalyeast.jpg | thumb | right | 150px | Gurpal's yeast transformants]] | ||
| + | |||
| + | Vicki is still in the process of getting more of the pADM743 plasmid from the original tube in case the current plasmid she is working with is contaminated. After transformation of pADM743 into <i>E.coli</i>, she yielded the following results: | ||
| + | *Positive control: TMTC (too many to count) | ||
| + | *Negative control: no colonies | ||
| + | *pADM743: TMTC | ||
| + | |||
| + | This shows promise, so she proceeded to set up overnight cultures for mini-prepping (with a control, of course). | ||
| + | |||
| + | After mini-prepping the pADM743, she ran a PCR on it using custom primers to check if (-)-limonene is present. Gel verification shows similar bands as before (4-5 bands per sample/lane). Vicki came to the conclusion that the plasmid that was given to her is <b>not</b> limonene. Because other team members are working with a variety of other synthases, she decided to dedicate the remainder of her summer to achieve the criteria for the gold medal: characterizing an existing part in the registry. She will take the limonene synthase (Part BBa_K118025 (Lims1)) from the biobrick registry and put that onto a yeast plasmid for characterization in yeast. At the same time, Daisy will be characterizing the same part in bacteria so that it can be compared against the data from yeast. | ||
| + | |||
| + | Vicki learned that sometimes, one needs to know when to quit and move onto a different aspect of the project such that it benefits the whole of team (especially when there is a time constraint). | ||
| + | |||
| + | ==IDI1, HMG2 metabolic genes== | ||
| + | |||
| + | Sam performed PCR on one sample of HMG2 plasmid and two samples of IDI1 plasmid to add an SMA1 cutsite upstream of the promoter on both genes. This PCR was unsuccessful; no bands were seen for any of the unpurified PCR product. | ||
| + | |||
| + | Gurpal performed mini plasmid preps, using the Invitrogen kit, one 2 overnight cultures: IDI1 and VEC plasmid. | ||
| + | |||
| + | ==ERG20 & erg20-2== | ||
| + | |||
| + | Gurpal performed a yeast transformation. He separately transformed pBS and pKS into wildtype yeast. It was a success! This is really awesome because now we can start to combine synthase genes and the ERG20/erg20-2 genes together and compare if more monoterpenes are produced! Yay, everyone gets a single colony! | ||
| + | |||
| + | [[File:ubcigemgurpalsam.jpg | thumb | left | 200px | Sam and Gurpal having fun in the lab]] | ||
| + | |||
| + | Gurpal also began to construct his ERG20 and erg20-2 biobricks. He did a PCR reaction to make many copies of these two genes. Jacob stores his PCR product in the 4°C fridge. Stay tuned next week... | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | ==Team Social at the Naam== | ||
| + | The Naam is a vegetarian restaurant located in Kitsilano (West side of the city of Vancouver). Despite the lack of meat, it was still very good! The dishes were huge and very filling. We learned a little more about each other and had a few laughs with each other, but especially at each other (Teasing each other is one our specialties!) Not yet satisfied, the team went to True Confections for dessert! | ||
| + | [[File:ubcigemnaam.jpg | frame | bottom | left | 200px]] | ||
| + | [[File:ubcigemnaam1.jpg | frame | bottom | right | 200px]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 23:14, 16 October 2011

 |
 |
 |
 |
 |
Contents |
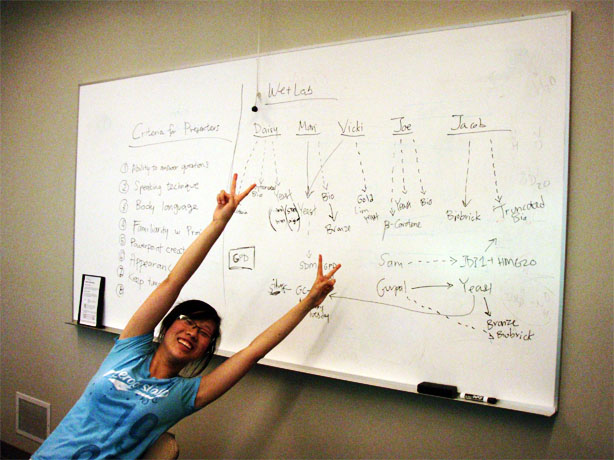
Lab Meeting - July 25
Not a lot happened at today's meeting.
Due to safety reasons, a reminder that everyone needs to be in pairs, especially after the typical working hours (9am-5pm).
Presentation
The team got together to discuss the presentation. We talked about what we liked and didn't like about the presentations we watched. For example, we didn't want all team members to be on the platform with the presenters because it feels distracting and unnecessary. Also, some of us thought that the flow of the story is very important; powerpoint slides shouldn't be consumed by wordy sentences but with clear and simple pictures with few words. In the end, we agreed that there would be 2-3 presenters.
We discussed the criteria to evaluate the potential presenters; the plan is for potential presenters to present a powerpoint on their role in the project, and feedback will be given at the end by everyone.
Constructing the Wiki
Vicki, Alina, and Gurpal started thinking of template ideas for the wiki. Because Vicki cannot draw or use photoshop to save her life, Alina and Gurpal volunteered to create most (if not all) of the images for the wiki. You know what this means: drawing contest... although Gurpal calls it, "Alina Wins!"
In the meantime, Vicki downloaded the trial version of Dreamweaver and began looking through the online tutorials. Her goal is to master Dreamweaver so that she can use it to design the format of the wiki. Let the late nights begin.
3-Carene
Daisy has been PCRing out the synthase with the yeast primers that contain the NotI and SmaI cut sites and an N-terminal His-tag. However, the forward primer is quite long and Daisy thinks there is some nonspecific annealing happening.
1,8-Cineole
The previous colonies of E. coli that should have had the 1,8-cineole synthase gene on pSB1C3 were shown to have different sized plasmids after overnight cultures and minipreps. Of the 5 colonies, 4 had plasmids a little over 2 kb in size, and one had a plasmid around 5 kb in size. The 1,8-cineole gene is 1.7 kb and the pSB1C3 plasmid that is left after the restriction digest is 2 kb, so the product should have been around 3.7 kb, suggestion it didn't work. A restriction digest with ECORI and PSTI showed only one band for the 5 kb product, and the 2 kb product showed bands at about 1.7 kb and 700 bp.
These results made no sense, so to check if there was a problem with the plasmid, pure pSB1C3 was run on an agarose gel. The gel showed a product well below the 500 bp mark on a 1 kb ladder.
This was rather disconcerting, so Jacob decided to run another biobrick PCR and try the ligation again with another source of the plasmid.
alpha-Pinene
Joe finally received his primers for PCR amplification of his SDM-alpha pinene synthase necessary for the ligation into yeast and biobrick plasmids. He did a PCR using these primers. However, no PCR products were seen after gel verification. The annealing temperature may have been too low at 55 degrees Celsius. Therefore, he needs to increase the annealing temperature to 60-72 degrees Celsius, the optimal temperature for Phusion Hot Start Polymerase.
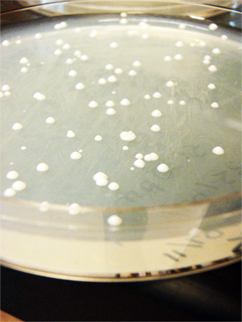
beta-Pinene
Marianne's transformations (yeast plasmids + synthase) show a few colonies each on 3 plates (original-his-GAL, SDM-no-his-GPD, SDM-no-his-GAL. Marianne conducted a colony PCR of these yeast plasmid and synthase transformants and gel verified them. The gel image shows bands at the correct lengths (~500bp), indicating that the ligations were successful! We have beta-pinene on yeast plasmids!
Marianne started on the Biobrick assembly; she restriction digested and ligated synthase and pSB1C3 backbone. Marianne set up an O/N culture for the yeast plasmids. Marianne transformed the Biobrick ligations and miniprepped the yeast plasmids. Marianne also transformed the miniprepped yeast plasmids(SND, SNL) into wildtype yeast, pKS yeast, pBS yeast.
(-)-Limonene
Vicki is still in the process of getting more of the pADM743 plasmid from the original tube in case the current plasmid she is working with is contaminated. After transformation of pADM743 into E.coli, she yielded the following results:
- Positive control: TMTC (too many to count)
- Negative control: no colonies
- pADM743: TMTC
This shows promise, so she proceeded to set up overnight cultures for mini-prepping (with a control, of course).
After mini-prepping the pADM743, she ran a PCR on it using custom primers to check if (-)-limonene is present. Gel verification shows similar bands as before (4-5 bands per sample/lane). Vicki came to the conclusion that the plasmid that was given to her is not limonene. Because other team members are working with a variety of other synthases, she decided to dedicate the remainder of her summer to achieve the criteria for the gold medal: characterizing an existing part in the registry. She will take the limonene synthase (Part BBa_K118025 (Lims1)) from the biobrick registry and put that onto a yeast plasmid for characterization in yeast. At the same time, Daisy will be characterizing the same part in bacteria so that it can be compared against the data from yeast.
Vicki learned that sometimes, one needs to know when to quit and move onto a different aspect of the project such that it benefits the whole of team (especially when there is a time constraint).
IDI1, HMG2 metabolic genes
Sam performed PCR on one sample of HMG2 plasmid and two samples of IDI1 plasmid to add an SMA1 cutsite upstream of the promoter on both genes. This PCR was unsuccessful; no bands were seen for any of the unpurified PCR product.
Gurpal performed mini plasmid preps, using the Invitrogen kit, one 2 overnight cultures: IDI1 and VEC plasmid.
ERG20 & erg20-2
Gurpal performed a yeast transformation. He separately transformed pBS and pKS into wildtype yeast. It was a success! This is really awesome because now we can start to combine synthase genes and the ERG20/erg20-2 genes together and compare if more monoterpenes are produced! Yay, everyone gets a single colony!
Gurpal also began to construct his ERG20 and erg20-2 biobricks. He did a PCR reaction to make many copies of these two genes. Jacob stores his PCR product in the 4°C fridge. Stay tuned next week...
Team Social at the Naam
The Naam is a vegetarian restaurant located in Kitsilano (West side of the city of Vancouver). Despite the lack of meat, it was still very good! The dishes were huge and very filling. We learned a little more about each other and had a few laughs with each other, but especially at each other (Teasing each other is one our specialties!) Not yet satisfied, the team went to True Confections for dessert!
 "
"