Team:Bielefeld-Germany/Labjournal
From 2011.igem.org
m (→Week 4: 23rd - 29th may) |
|||
| (28 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
'''Labjournal:''' On this page we summarize the (successful) results and achievements of our teamwork. | '''Labjournal:''' On this page we summarize the (successful) results and achievements of our teamwork. | ||
| - | + | ==Week 1: 2nd - 8th May== | |
| - | ==Week 1: 2nd - 8th | + | |
Bisphenol A: | Bisphenol A: | ||
* cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic) | * cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic) | ||
| - | * cloning of | + | * cloning of fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter |
* testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | * testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | ||
* expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] | * expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] | ||
S-layer: | S-layer: | ||
* successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html ''Corynebacterium glutamicum''] and ''Corynebacterium crenatum'' | * successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html ''Corynebacterium glutamicum''] and ''Corynebacterium crenatum'' | ||
| - | * successful cloning of the S-layer gene of '' | + | * successful cloning of the S-layer gene of ''Brevibacterium flavum'' without TAT-sequence |
Organizational: | Organizational: | ||
* meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld | * meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld | ||
| - | + | ==Week 2: 9th - 15th May== | |
| - | ==Week 2: 9th - 15th | + | |
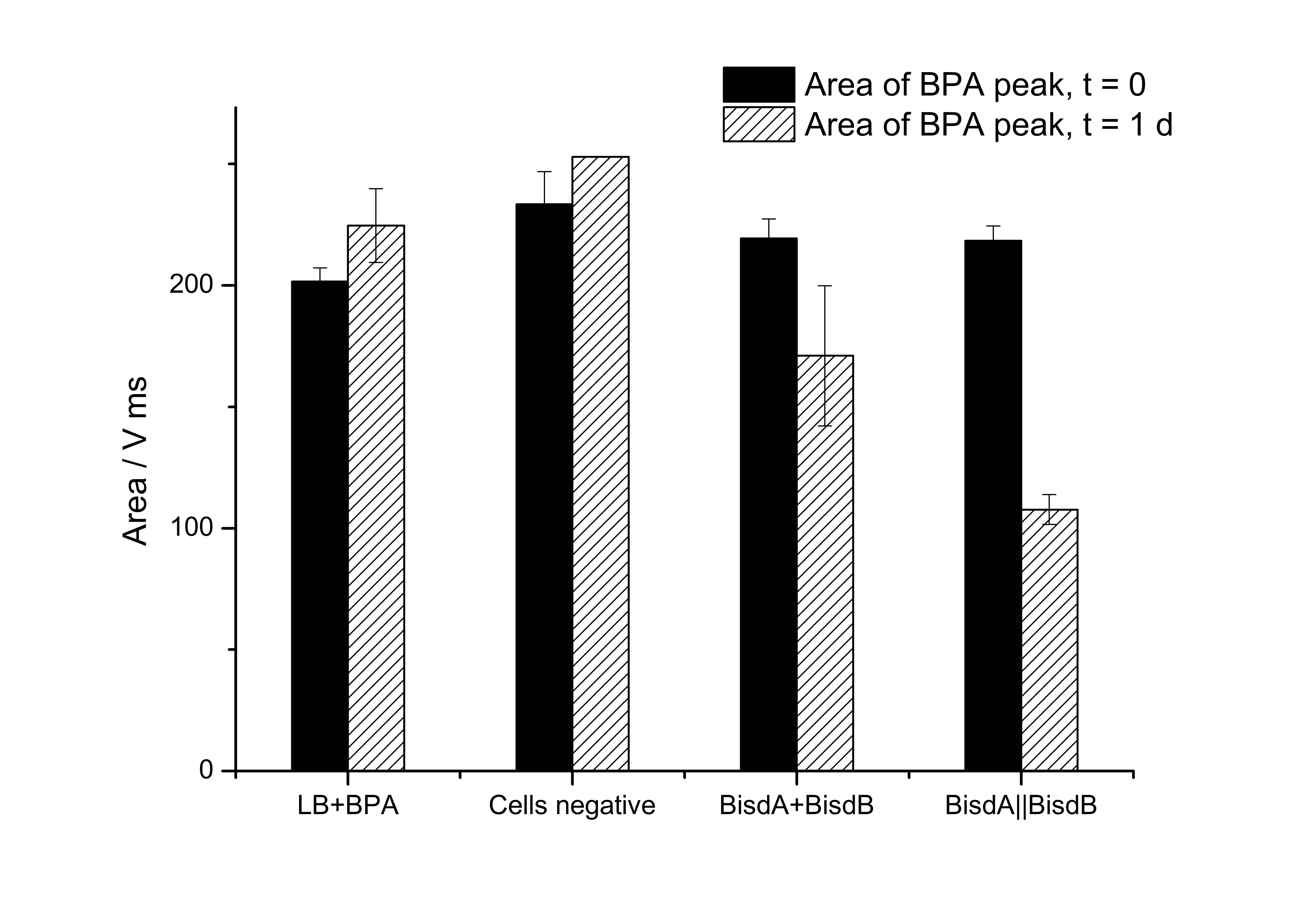
[[Image:Bielefeld2011_BPAAbbau_Versuch_1.jpg|300px|thumb|right| '''Figure 1: HPLC results of the first experiment on BPA degradation in ''E. coli'' TOP10. Cultivations were carried out in LB medium with 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein. ''']] | [[Image:Bielefeld2011_BPAAbbau_Versuch_1.jpg|300px|thumb|right| '''Figure 1: HPLC results of the first experiment on BPA degradation in ''E. coli'' TOP10. Cultivations were carried out in LB medium with 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein. ''']] | ||
Bisphenol A: | Bisphenol A: | ||
* assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system) | * assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system) | ||
| - | * HPLC results: Fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare | + | * HPLC results: Fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare Figure 1) |
S-layer: | S-layer: | ||
* expression of the S-layer gene without TAT-sequence of ''C. glutamicum'' in ''E. coli'' [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results | * expression of the S-layer gene without TAT-sequence of ''C. glutamicum'' in ''E. coli'' [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results | ||
| Line 30: | Line 28: | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | ==Week 3: 16th - 22nd | + | ==Week 3: 16th - 22nd May== |
Bisphenol A: | Bisphenol A: | ||
* successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP<sup>+</sup> oxidoreductase] gene from ''E. coli'' TOP10 | * successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP<sup>+</sup> oxidoreductase] gene from ''E. coli'' TOP10 | ||
| Line 37: | Line 35: | ||
* successful cloning of the S-layer genes of ''B. flavum'' and ''C. halotolerans'' without TAT-sequence, without lipid anchor and without both (only self-assembly domain) | * successful cloning of the S-layer genes of ''B. flavum'' and ''C. halotolerans'' without TAT-sequence, without lipid anchor and without both (only self-assembly domain) | ||
| - | + | ==Week 4: 23rd - 29th May== | |
| - | ==Week 4: 23rd - 29th | + | |
Bisphenol A: | Bisphenol A: | ||
* establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS) | * establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS) | ||
| Line 45: | Line 42: | ||
* substantiating our contribution to the GENIALE | * substantiating our contribution to the GENIALE | ||
| - | ==Week 5: 30th | + | ==Week 5: 30th May - 5th June== |
Bisphenol A: | Bisphenol A: | ||
* beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>) | * beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>) | ||
Organizational: | Organizational: | ||
| - | * meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in | + | * meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in July |
| - | + | ==Week 6: 6th - 12th June== | |
| - | ==Week 6: 6th - 12th | + | |
Organizational: | Organizational: | ||
* presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention] | * presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention] | ||
| - | + | ==Week 7: 13th - 19th June== | |
| - | ==Week 7: 13th - 19th | + | S-layer: |
| - | S- | + | * successful fusion of modified ''cspB'' genes of [http://hamap.expasy.org/proteomes/CORGL.html ''C. glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] with a monomeric RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. |
| - | * successful fusion of modified cspB genes of [http://hamap.expasy.org/proteomes/CORGL.html ''C. glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] with a monomeric RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | + | |
* ''C.glutamicum'': | * ''C.glutamicum'': | ||
| - | ** K525131: fusion of cspB including lipid anchor and TAT-sequence | + | ** K525131: fusion of ''cspB'' including lipid anchor and TAT-sequence <partinfo>K525121</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | ** K525133: fusion of cspB including lipid anchor without TAT-sequence | + | ** K525133: fusion of ''cspB'' including lipid anchor without TAT-sequence <partinfo>K525123</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
* ''C. halotolerans'': | * ''C. halotolerans'': | ||
| - | ** K525233: fusion of cspB including lipid anchor without TAT-sequence | + | ** K525233: fusion of ''cspB'' including lipid anchor without TAT-sequence <partinfo>K525223</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | ** K525224: fusion of cspB including TAT-sequence without lipid anchor | + | ** K525224: fusion of ''cspB'' including TAT-sequence without lipid anchor <partinfo>K525224</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | * First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD<sub>600</sub>) but in a higher fluorescence level | + | * First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD<sub>600</sub>) but in a higher fluorescence level. The variations of the induction time point has no significant influence on growth and the progression of fluorescence. |
[[Image:Bielefeld2011 Kult 19.06 OD600.png|450px|thumb|left| '''Figure 2: Growth curve of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] | [[Image:Bielefeld2011 Kult 19.06 OD600.png|450px|thumb|left| '''Figure 2: Growth curve of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] | ||
| - | [[Image:Bielefeld2011_Kult_1906_RFU.png|450px|thumb|right|'''Figure 3: Fluorescence | + | [[Image:Bielefeld2011_Kult_1906_RFU.png|450px|thumb|right|'''Figure 3: Fluorescence development of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 during cultivation using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] |
Bisphenol A: | Bisphenol A: | ||
* cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites | * cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites | ||
| - | + | ==Week 8: 20th - 26th June== | |
| - | ==Week 8: 20th - 26th | + | |
S-layer: | S-layer: | ||
* successful cloning of K525232 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | * successful cloning of K525232 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | ||
| - | ** K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] without lipid anchor and TAT-sequence | + | ** K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] without lipid anchor and TAT-sequence <partinfo>K525222</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]. |
Organizational: | Organizational: | ||
* finishing our first press release | * finishing our first press release | ||
* all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived | * all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived | ||
| - | + | ==Week 9: 27th June - 3rd July== | |
| - | ==Week 9: 27th | + | |
EXAMS ! | EXAMS ! | ||
| - | + | ==Week 10: 4th July - 10th July== | |
| - | ==Week 10: 4th | + | |
Bisphenol A: | Bisphenol A: | ||
| - | * experiments on the influence of temperature, | + | * experiments on the influence of temperature, promoter strength and the characteristics of the fusion protein <partinfo>K123000</partinfo> || <partinfo>K123001</partinfo> on BPA degradation |
| - | S- | + | S-layer: |
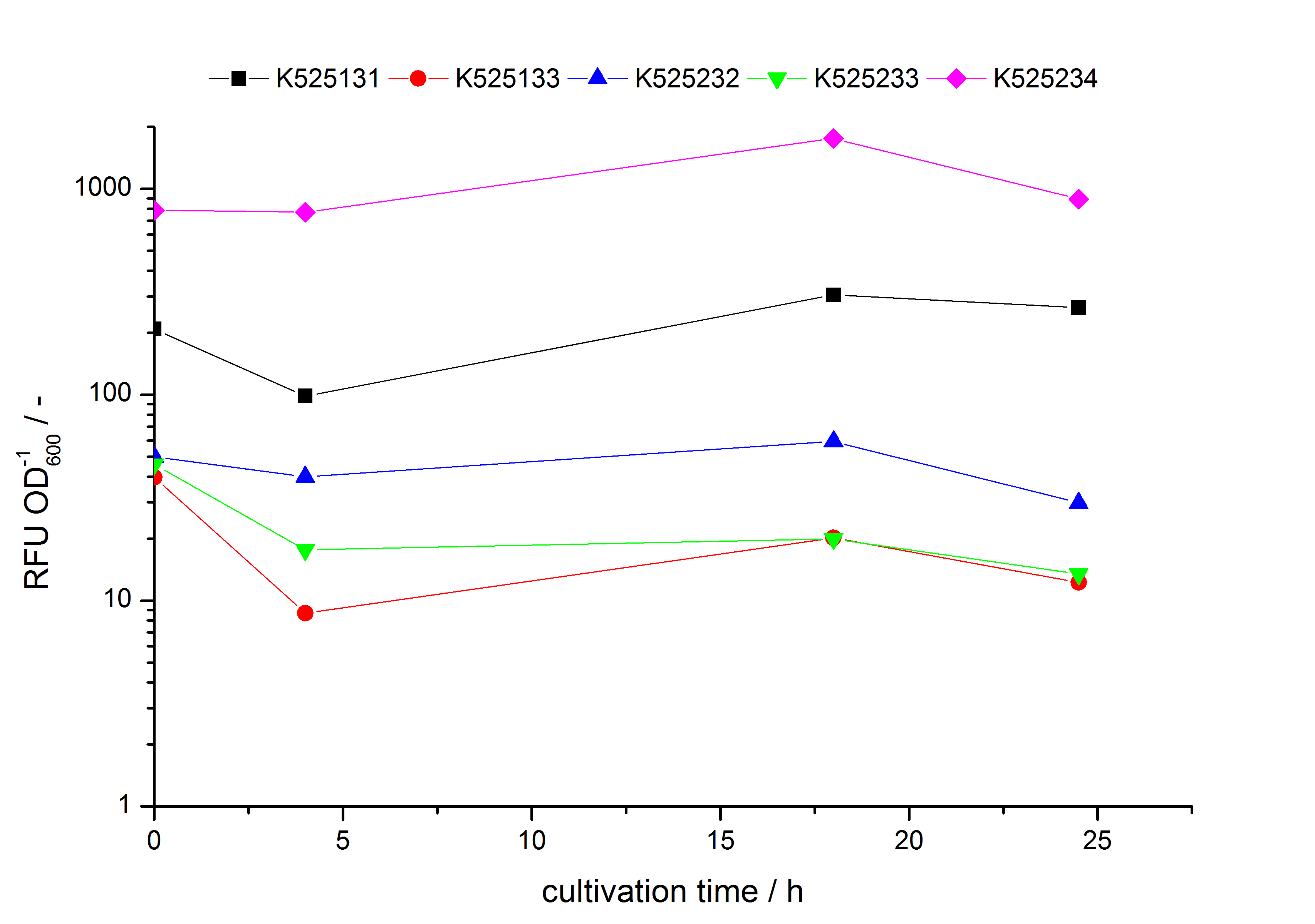
| - | * Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in ''E. coli'' KRX to test the functional efficiency of Corynebacterium TAT-sequence in ''E.coli'' and the effect of the autoinduction protocol. After one day of cultivation | + | * Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in ''E. coli'' KRX to test the functional efficiency of ''Corynebacterium'' TAT-sequence in ''E.coli'' and the effect of the autoinduction protocol. After one day of cultivation, an increased fluorescence in all periplasm fractions of ''E. coli'' expressing S-layer constructs with TAT-sequence could be recognized. This is an indication that the ''Corynebacterium'' TAT-signal sequence is functional in ''E. coli''. Comparing the manual and the autoinduction protocol, no significant changes in fluorescence (after 14 h) are identifiable. |
[[Image:Bielefeld2011 Kult 07.07 RFU pellet.png|450px|thumb|left| '''Figure 4: Relative fluorescence of the S-layer/mRFP fusion proteins in the cell suspension less the distinct fluorescence of ''E. coli'' KRX. All cultivations were carried out in LB medium at 37 ˚C except K525131<sub>Glc</sub> which was in autoinduction medium. The expression of the S-layers in LB medium were induced adding 0,1 % L-rhamnose at OD<sub>600</sub> = 0,6.''']] | [[Image:Bielefeld2011 Kult 07.07 RFU pellet.png|450px|thumb|left| '''Figure 4: Relative fluorescence of the S-layer/mRFP fusion proteins in the cell suspension less the distinct fluorescence of ''E. coli'' KRX. All cultivations were carried out in LB medium at 37 ˚C except K525131<sub>Glc</sub> which was in autoinduction medium. The expression of the S-layers in LB medium were induced adding 0,1 % L-rhamnose at OD<sub>600</sub> = 0,6.''']] | ||
| Line 100: | Line 92: | ||
* presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research). | * presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research). | ||
| - | + | ==Week 11: 11th July - 17th July== | |
| - | ==Week 11: 11th | + | Bisphenol A / S-layer: |
| - | Bisphenol A / S- | + | * our BioBrick order (some fluorescent proteins and cleavage sites) from iGEM HQ arrived |
| - | * our BioBrick order (some | + | |
S-layer: | S-layer: | ||
| - | * our synthesized S- | + | * our synthesized S-layers SgsE and SbpA finally arrived |
Organizational: | Organizational: | ||
* presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos]) | * presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos]) | ||
| - | + | ==Week 12: 18th July - 24th July== | |
| - | ==Week 12: 18th | + | |
Organizational: | Organizational: | ||
* presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld | * presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld | ||
| - | * meeting | + | * meeting with the iGEM teams from Delft/NL, Edinburgh/UK, Odense/DK. Freiburg/DE and Ljubljana/SL at the Symposium ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627300144576/ Photos]) was fun! |
Bisphenol A / S-Layer: | Bisphenol A / S-Layer: | ||
| - | * removing illegal | + | * removing illegal restriction sites to get valid BioBricks |
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
* cloning the NAD<sup>+</sup>-dependent DNA ligase from ''E. coli'' into BioBrick backbones | * cloning the NAD<sup>+</sup>-dependent DNA ligase from ''E. coli'' into BioBrick backbones | ||
| - | + | ==Week 13: 25th July - 31th July== | |
| - | ==Week 13: 25th | + | S-layer: |
| - | S- | + | |
* fusing the synthesized S-Layers to a bunch of fluorescent proteins | * fusing the synthesized S-Layers to a bunch of fluorescent proteins | ||
* successful cloning of K525231 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | * successful cloning of K525231 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | ||
| - | ** K525231: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] including lipid anchor and TAT-sequence | + | ** K525231: fusion of modified ''cspB'' of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] including lipid anchor and TAT-sequence <partinfo>K525221</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]. |
| - | * testing the basal transcription of ''E. coli'' KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD<sub>600</sub> reveals the basal transcription of ''E. coli'' KRX. L-rhamnose in the medium leads | + | * testing the basal transcription of ''E. coli'' KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD<sub>600</sub> reveals the basal transcription of ''E. coli'' KRX. L-rhamnose in the medium leads to a ten times higher RFU per OD<sub>600</sub>. |
[[Image:Bielefeld2011 Kult 29.07 BF OD.png|450px|thumb|left| '''Figure 6: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C. glutamicum'']. Cultivations were carried out in autoinduction medium at 37 ˚C with and without adding inducer (L-rhamnose). The induced cultivations are marked with index I and the uninduced are marked with index U.''']] | [[Image:Bielefeld2011 Kult 29.07 BF OD.png|450px|thumb|left| '''Figure 6: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C. glutamicum'']. Cultivations were carried out in autoinduction medium at 37 ˚C with and without adding inducer (L-rhamnose). The induced cultivations are marked with index I and the uninduced are marked with index U.''']] | ||
| Line 140: | Line 129: | ||
* testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F) | * testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F) | ||
| - | + | ==Week 14: 1st August - 7th August== | |
| - | ==Week 14: 1st | + | |
Bisphenol A: | Bisphenol A: | ||
* BPA analysis with [[Team:Bielefeld-Germany/Protocols#Extraction_with_ethylacetate | extraction]] and LC-MS finally works and is very accurate | * BPA analysis with [[Team:Bielefeld-Germany/Protocols#Extraction_with_ethylacetate | extraction]] and LC-MS finally works and is very accurate | ||
| Line 180: | Line 168: | ||
|} | |} | ||
| - | + | ==Week 15: 8th August - 14th August== | |
| - | ==Week 15: 8th | + | |
Bisphenol A: | Bisphenol A: | ||
| - | * BPA analysis with [[Team:Bielefeld-Germany/Protocols/Analytics#Extraction_with_ethylacetate | extraction]] and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare | + | * BPA analysis with [[Team:Bielefeld-Germany/Protocols/Analytics#Extraction_with_ethylacetate | extraction]] and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare Figure 25) |
* measuring of more samples from cultivations with BPA degrading BioBricks for further characterization | * measuring of more samples from cultivations with BPA degrading BioBricks for further characterization | ||
* we discovered some interesting results in our MS data - soon more | * we discovered some interesting results in our MS data - soon more | ||
* testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins | * testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins | ||
* testing the influence of BPA on the growth of ''E. coli'' | * testing the influence of BPA on the growth of ''E. coli'' | ||
| - | * developing a | + | * developing a model for BPA degradation by ''E. coli'' (compare Figure 26) |
[[Image:iGEM_Bielefeld_Vergleich_Analysemethoden_BPA.jpg|450px|thumb|left| '''Figure 25: HPLC results and optical density of experiments on BPA degradation of ''E. coli'' KRX. Cultivations were carried out in LB medium with ~ 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken every three hours over one day. BPA concentration was measured by HPLC with either UV detector or ESI-qTOF-MS. 502: negative control (<partinfo>K123000</partinfo>), 512: polycistronic <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, 517: fusionprotein between <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, every part behind medium strong constitutive promoter. ''']] | [[Image:iGEM_Bielefeld_Vergleich_Analysemethoden_BPA.jpg|450px|thumb|left| '''Figure 25: HPLC results and optical density of experiments on BPA degradation of ''E. coli'' KRX. Cultivations were carried out in LB medium with ~ 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken every three hours over one day. BPA concentration was measured by HPLC with either UV detector or ESI-qTOF-MS. 502: negative control (<partinfo>K123000</partinfo>), 512: polycistronic <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, 517: fusionprotein between <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, every part behind medium strong constitutive promoter. ''']] | ||
| Line 197: | Line 184: | ||
* MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by ''cspB'' gene) in ''E. coli'' KRX. | * MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by ''cspB'' gene) in ''E. coli'' KRX. | ||
| - | + | ==Week 16: 15th August - 21st August== | |
| - | ==Week 16: 15th | + | |
S-layer: | S-layer: | ||
* finally found our S-layer proteins in ''E. coli'' by analyzing the range of sizes in the polyacrylamid with a MALDI-TOF method-> now we can plan purification strategy | * finally found our S-layer proteins in ''E. coli'' by analyzing the range of sizes in the polyacrylamid with a MALDI-TOF method-> now we can plan purification strategy | ||
| Line 208: | Line 194: | ||
** seems like they can't | ** seems like they can't | ||
| - | + | ==Week 17: 22nd August - 28th August== | |
| - | ==Week 17: 22nd | + | |
S-layer: | S-layer: | ||
* cultivation and test purification of ''SgsE'' fusion proteins (<partinfo>K525304</partinfo>, <partinfo>K525305</partinfo> and <partinfo>K525306</partinfo>) | * cultivation and test purification of ''SgsE'' fusion proteins (<partinfo>K525304</partinfo>, <partinfo>K525305</partinfo> and <partinfo>K525306</partinfo>) | ||
| Line 218: | Line 203: | ||
* cloning of fusion protein FNR:BisdA:BisdB successful | * cloning of fusion protein FNR:BisdA:BisdB successful | ||
| - | + | ==Week 18: 29th August - 4th September== | |
| - | ==Week 18: 29th | + | |
S-layer: | S-layer: | ||
* developing IEX clean-up for S-layers from ''Corynebacterium halotolerans'' | * developing IEX clean-up for S-layers from ''Corynebacterium halotolerans'' | ||
| Line 230: | Line 214: | ||
* participating at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld with a [http://www.geniale-bielefeld.de/programm/detail/date/456/show/Event/ lab tour] and a [http://www.geniale-bielefeld.de/programm/detail/date/295/show/Event/ presentation with public discussion afterwards (science café)]. Our participation is also mentioned in the regional newspaper [http://www.westfalen-blatt.de/nachricht/2011-08-30-wie-ein-hamster-im-laufrad/?cHash=b8421d55eb00ebf8532ebfa677a7e18f Westfalen Blatt] and in the internet on [http://www.direkt-bielefeld.de/Wissenschaft-im-Doppelpack-108856.html Bielefeld Direkt]. | * participating at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld with a [http://www.geniale-bielefeld.de/programm/detail/date/456/show/Event/ lab tour] and a [http://www.geniale-bielefeld.de/programm/detail/date/295/show/Event/ presentation with public discussion afterwards (science café)]. Our participation is also mentioned in the regional newspaper [http://www.westfalen-blatt.de/nachricht/2011-08-30-wie-ein-hamster-im-laufrad/?cHash=b8421d55eb00ebf8532ebfa677a7e18f Westfalen Blatt] and in the internet on [http://www.direkt-bielefeld.de/Wissenschaft-im-Doppelpack-108856.html Bielefeld Direkt]. | ||
| - | + | ==Week 19: 5th September - 11th September== | |
| - | ==Week 19: 5th | + | |
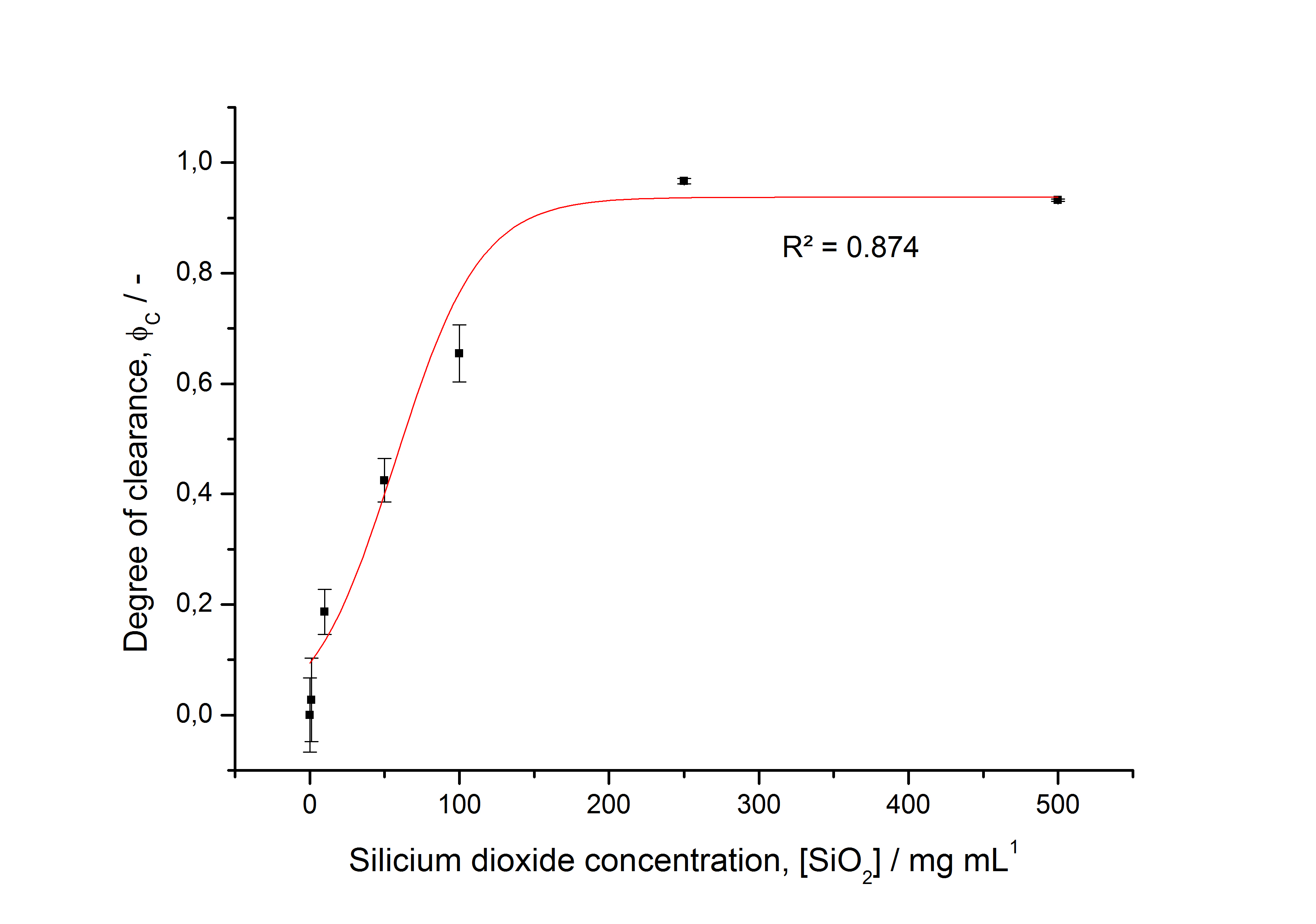
[[Image:Bielefeld-Germany2011-saturationdegree305.jpg|500px|thumb|'''Fig. 27: Immobilization behaviour of fluorescent S-layer fusion protein SgsE :: mCitrine as a function of silica bead concentration. Data fitted with Michaelis-Menten function. ''']] | [[Image:Bielefeld-Germany2011-saturationdegree305.jpg|500px|thumb|'''Fig. 27: Immobilization behaviour of fluorescent S-layer fusion protein SgsE :: mCitrine as a function of silica bead concentration. Data fitted with Michaelis-Menten function. ''']] | ||
S-layer: | S-layer: | ||
* coating silica beads with fluorescent S-layer fusion protein SgsE | mCitrine, remove these proteins again and find them in SDS-PAGE | * coating silica beads with fluorescent S-layer fusion protein SgsE | mCitrine, remove these proteins again and find them in SDS-PAGE | ||
| - | * characterizing immobilization behaviour of S-layers on silica beads -> seems to work ( | + | * characterizing immobilization behaviour of S-layers on silica beads -> seems to work (Figure 27) |
* cultivate bigger amounts of and develop new purification strategy for S-layer fusion proteins | * cultivate bigger amounts of and develop new purification strategy for S-layer fusion proteins | ||
* testing purification of S-layer fusion proteins with his-tag | * testing purification of S-layer fusion proteins with his-tag | ||
| Line 241: | Line 224: | ||
* also SbpA | mCitrine fusion protein | * also SbpA | mCitrine fusion protein | ||
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
| - | * | + | * Suitability of autonomously produced DNA ligase (<partinfo>K525710</partinfo>) from ''E. coli'' for the NAD<sup>+</sup> bioassay could be demonstrated after treatment with NMN for deadenylation. Checking out the limit of detection right now. |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | + | ==Week 20: 12th September - 18th September== | |
| - | ==Week 20: 12th | + | |
S-layer: | S-layer: | ||
* testing a purification and coating protocol for S-layer fusion protein mCitrine|SbpA. | * testing a purification and coating protocol for S-layer fusion protein mCitrine|SbpA. | ||
| Line 256: | Line 238: | ||
* NAD<sup>+</sup> assay works :) | * NAD<sup>+</sup> assay works :) | ||
| - | + | ==Week 21: 19th September - 25th September== | |
| - | ==Week 21: 19th | + | |
Organizational: | Organizational: | ||
| - | * lab work is finished, the last measurements and | + | * lab work is finished, the last measurements and cloning experiments into pSB1C3 are done |
* now it is time to fill the Wiki and Partsregistry with brilliant data until European Wikifreeze @ September 21st 11:59 pm EDT! | * now it is time to fill the Wiki and Partsregistry with brilliant data until European Wikifreeze @ September 21st 11:59 pm EDT! | ||
| - | + | ==Week 22: 26th September - 2nd October== | |
| - | ==Week 22: 26th | + | Preparation of the presentation and the poster for the European Regional Jamboree. |
| - | Preparation of the presentation and the poster for the | + | |
* practicing the presentation with the Coryne and fermentation engineering working group as audience. | * practicing the presentation with the Coryne and fermentation engineering working group as audience. | ||
| - | We are qualified for the final in Boston!!! Now labwork can continue. | + | '''We are qualified for the final in Boston!!! Now labwork can continue.''' |
| - | + | ||
| - | ==Week 23: 3rd | + | ==Week 23: 3rd October - 9th October== |
[[Image:Bielefeld-Germany-2011_Selectivity-of-LigA.png|450px|thumb|right| '''Figure 28: LigA shows high selectivity for NAD<sup>+</sup>.''' The final concentration of all analytes was 100 nM. The responses were evaluated on the basis of the average fluorescence enhancement rate in a range of 200 s after addition of each analyte into the NAD<sup>+</sup> bioassay. The dotted line marks the threshold indicating the intensity of background signal. All data are normalized to the NAD<sup>+</sup> value.]] | [[Image:Bielefeld-Germany-2011_Selectivity-of-LigA.png|450px|thumb|right| '''Figure 28: LigA shows high selectivity for NAD<sup>+</sup>.''' The final concentration of all analytes was 100 nM. The responses were evaluated on the basis of the average fluorescence enhancement rate in a range of 200 s after addition of each analyte into the NAD<sup>+</sup> bioassay. The dotted line marks the threshold indicating the intensity of background signal. All data are normalized to the NAD<sup>+</sup> value.]] | ||
S-layer: | S-layer: | ||
| - | *scale-up cultivation of ''E. coli'' expressing [http://partsregistry.org/Part:BBa_K525305 | + | *scale-up cultivation of ''E. coli'' expressing [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] fusion protein. |
*[https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Fusion_proteins_of_SgsE_and_SbpA inclusion body purification] and testing further purification strategies. | *[https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Fusion_proteins_of_SgsE_and_SbpA inclusion body purification] and testing further purification strategies. | ||
| - | *assembly of [http://partsregistry.org/Part:BBa_K525422 K525422] containing | + | *assembly of [http://partsregistry.org/Part:BBa_K525422 K525422] containing mCitrine|SbpA fused with a His-tag ([http://partsregistry.org/Part:BBa_K157011 K157011]) for easer purification. |
*assembly of [http://partsregistry.org/Part:BBa_K525311 K525311] consisting of the SbpA S-layer fused with firefly-luciferase ([http://partsregistry.org/Part:BBa_K525999 K525999]) for testing an enzyme reaction on immobilized S-layer proteins. | *assembly of [http://partsregistry.org/Part:BBa_K525311 K525311] consisting of the SbpA S-layer fused with firefly-luciferase ([http://partsregistry.org/Part:BBa_K525999 K525999]) for testing an enzyme reaction on immobilized S-layer proteins. | ||
*ordering silicon wafer as a new surface for immobilization. | *ordering silicon wafer as a new surface for immobilization. | ||
| - | * | + | *successful cloning of fusion protein of K525303 and K525560 [http://partsregistry.org/Part:BBa_K525001 (K525001)],[http://partsregistry.org/Part:BBa_K525311 (K525311)], [http://partsregistry.org/Part:BBa_K525006 SbpA|LigA (K525006)], [http://partsregistry.org/Part:BBa_K525422 (K525422)],[http://partsregistry.org/Part:BBa_K525322 (K525322)] |
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
* testing selectivity of LigA (<partinfo>BBa_K525710</partinfo>) for its substrate NAD<sup>+</sup> to verify further applications of the molecular beacon based NAD<sup>+</sup> bioassay dealing with NAD<sup>+</sup> detection in analyte mixtures such as cell lysates. | * testing selectivity of LigA (<partinfo>BBa_K525710</partinfo>) for its substrate NAD<sup>+</sup> to verify further applications of the molecular beacon based NAD<sup>+</sup> bioassay dealing with NAD<sup>+</sup> detection in analyte mixtures such as cell lysates. | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | ==Week 24: 10th | + | ==Week 24: 10th October - 16th October== |
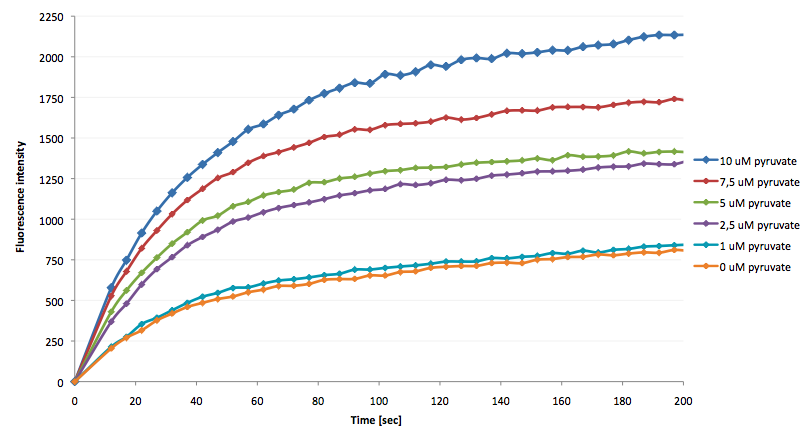
[[Image:Bielefeld-Germany-2011 Fluorescence-enhancement-after addition-of-LDH-reaction-mix.png|450px|thumb|right| '''Figure 29: Fluorescence enhancement rate after addition of LDH reaction mix with various pyruvate concentrations into a LigA-dependent NAD<sup>+</sup> bioassay.''']] | [[Image:Bielefeld-Germany-2011 Fluorescence-enhancement-after addition-of-LDH-reaction-mix.png|450px|thumb|right| '''Figure 29: Fluorescence enhancement rate after addition of LDH reaction mix with various pyruvate concentrations into a LigA-dependent NAD<sup>+</sup> bioassay.''']] | ||
| Line 292: | Line 271: | ||
*new inclusion body purification of K525305 and K525405 to get new biological material to test the further purification methods. | *new inclusion body purification of K525305 and K525405 to get new biological material to test the further purification methods. | ||
*establishment of ion exchange chromatography (IEX) and hydrophobic interaction chromatography (HIC) as futher purification steps for S-layer proteins from ''Lysinibacillus sphaericus'' and ''Geobacillus stearothermophilus''. | *establishment of ion exchange chromatography (IEX) and hydrophobic interaction chromatography (HIC) as futher purification steps for S-layer proteins from ''Lysinibacillus sphaericus'' and ''Geobacillus stearothermophilus''. | ||
| - | **diethylaminoethyl cellulose (DEAE) as IEX and butyl sepharose™ 4 Fast Flow as HIC | + | **diethylaminoethyl cellulose (DEAE) as IEX and butyl sepharose™ 4 Fast Flow as HIC purification media. |
| - | *test and scale-up expression of [http://partsregistry.org/Part:BBa_K525322 | + | *test and scale-up expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] in ''E. coli'' KRX. |
| - | **after enzymatic cell lysis with lysozyme and inclusion body purification highest luminescence was | + | **after enzymatic cell lysis with lysozyme and inclusion body purification highest luminescence was measured in the lysis supernatant. This implicates that the fusion protein does not form inclusion bodies. |
| - | * | + | *successful cloning of fusion protein of CspB and luciferase with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Genetics Gibson assembly] |
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
*the NAD<sup>+</sup> bioassay was successfully coupled to the NADH-dependent reaction of lactic acid dehydrogenase and could therefore be used to quantify pyruvate | *the NAD<sup>+</sup> bioassay was successfully coupled to the NADH-dependent reaction of lactic acid dehydrogenase and could therefore be used to quantify pyruvate | ||
BPA-degradation: | BPA-degradation: | ||
| - | * | + | *successful cloning of [http://partsregistry.org/Part:BBa_K525562 (K525562)] ( Fusion protein of [http://partsregistry.org/Part:BBa_K525499 (K525499)], [http://partsregistry.org/Part:BBa_K123000 (K123000)]and [http://partsregistry.org/Part:BBa_K123001 (K5123001)] with middle strong promoter) and [http://partsregistry.org/Part:BBa_K525534 (K525534)] |
| - | ==Week 25: 17th | + | ==Week 25: 17th October - 23th October== |
S-layer: | S-layer: | ||
| - | *new expression of K525311 to develop an individual purification strategy without inclusion body purification. | + | *new expression of <partinfo>K525311</partinfo> to develop an individual purification strategy without inclusion body purification. |
| - | *testing a new purification | + | *testing a new purification strategy for the fusion protein of the SgsE and the firefly-luciferase based on the IEX and HIC |
| - | *first expression of [http://partsregistry.org/Part:BBa_K525322 | + | *first expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] and successful His-tag affinity chromatography based purification using denaturing conditions. |
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
*testing LigA for molecular cloning | *testing LigA for molecular cloning | ||
| - | + | ==Week 26: 24th October - 30th October== | |
| - | ==Week 26: 24th | + | |
S-layer: | S-layer: | ||
*test expression of [http://partsregistry.org/Part:BBa_K525534 reductase|His-tag (K525534)] in ''E. coli'' KRX. and testing non denaturating His-tag affinity chromatography. | *test expression of [http://partsregistry.org/Part:BBa_K525534 reductase|His-tag (K525534)] in ''E. coli'' KRX. and testing non denaturating His-tag affinity chromatography. | ||
| - | *recrystallization of [http://partsregistry.org/Part:BBa_K525305 | + | *recrystallization of [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] on silicon wafer using two different immobilization strategies. |
*trying to get pictures of the S-layer nanostructures on coated silicon wafers using reflection electron microscope (REM) and atomic force microscopy (AFM) | *trying to get pictures of the S-layer nanostructures on coated silicon wafers using reflection electron microscope (REM) and atomic force microscopy (AFM) | ||
| - | ** | + | **not enough time left to find a successful method |
Latest revision as of 23:09, 28 October 2011


Labjournal: On this page we summarize the (successful) results and achievements of our teamwork.
Week 1: 2nd - 8th May
Bisphenol A:
- cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic)
- cloning of fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter
- testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection
- expression of the successfully assembled BPA degrading BioBricks in E. coli [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10]
S-layer:
- successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html Corynebacterium glutamicum] and Corynebacterium crenatum
- successful cloning of the S-layer gene of Brevibacterium flavum without TAT-sequence
Organizational:
- meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld
Week 2: 9th - 15th May
Bisphenol A:
- assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system)
- HPLC results: Fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare Figure 1)
S-layer:
- expression of the S-layer gene without TAT-sequence of C. glutamicum in E. coli [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results
- successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 Corynebacterium halotolerans]
Organizational:
- moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec]
Week 3: 16th - 22nd May
Bisphenol A:
- successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP+ oxidoreductase] gene from E. coli TOP10
S-layer:
- successful cloning of the complete S-layer gene cspB of C. glutamicum, C. crenatum and C. halotolerans
- successful cloning of the S-layer genes of B. flavum and C. halotolerans without TAT-sequence, without lipid anchor and without both (only self-assembly domain)
Week 4: 23rd - 29th May
Bisphenol A:
- establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS)
Organizational:
- arrange a BBQ for our workgroup in the CeBiTec to get to know our co-workers
- substantiating our contribution to the GENIALE
Week 5: 30th May - 5th June
Bisphenol A:
- beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>)
Organizational:
- meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in July
Week 6: 6th - 12th June
Organizational:
- presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention]
Week 7: 13th - 19th June
S-layer:
- successful fusion of modified cspB genes of [http://hamap.expasy.org/proteomes/CORGL.html C. glutamicum] and [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] with a monomeric RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) using Gibson assembly.
- C.glutamicum:
- K525131: fusion of cspB including lipid anchor and TAT-sequence <partinfo>K525121</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- K525133: fusion of cspB including lipid anchor without TAT-sequence <partinfo>K525123</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- C. halotolerans:
- K525233: fusion of cspB including lipid anchor without TAT-sequence <partinfo>K525223</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- K525224: fusion of cspB including TAT-sequence without lipid anchor <partinfo>K525224</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD600) but in a higher fluorescence level. The variations of the induction time point has no significant influence on growth and the progression of fluorescence.
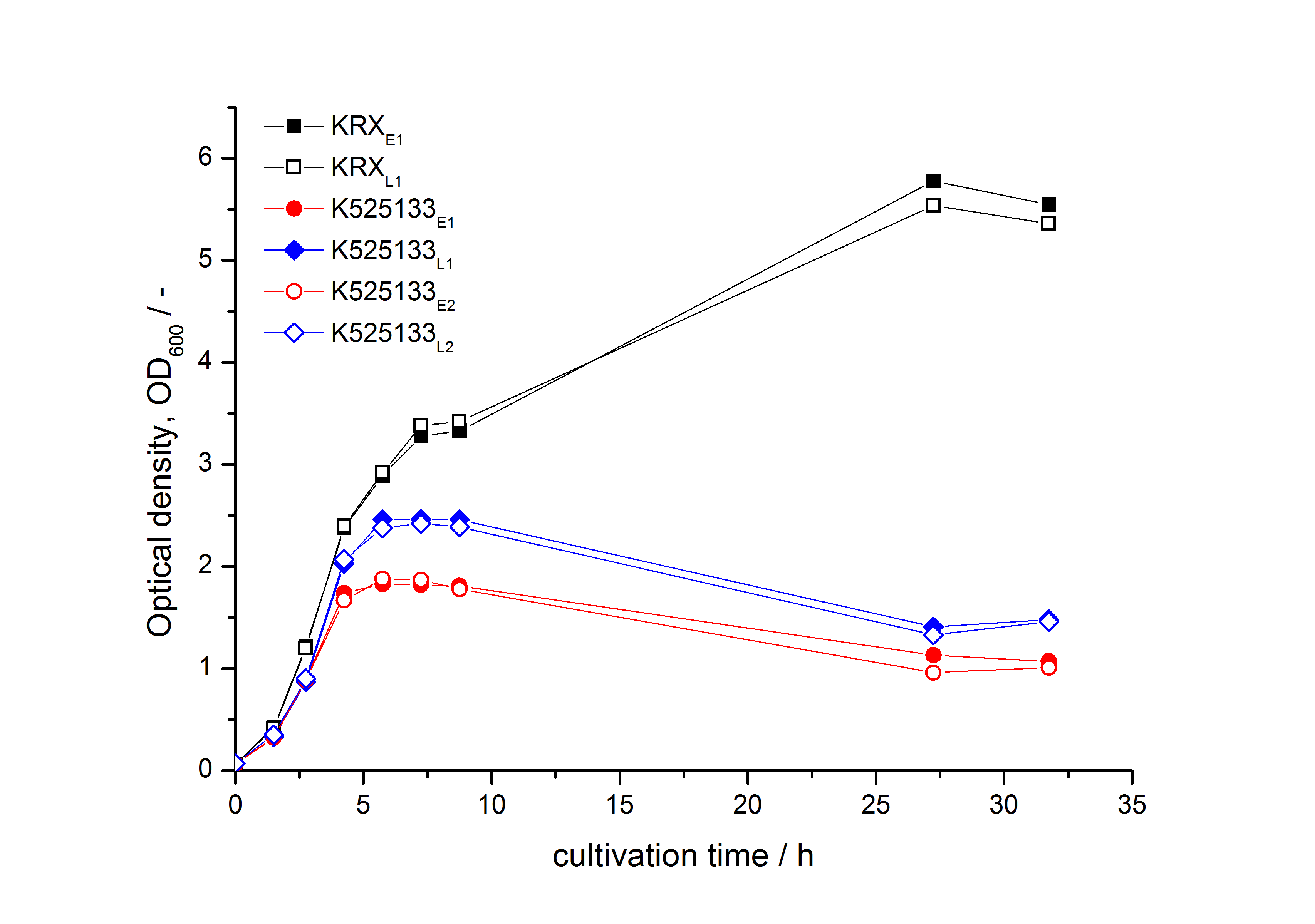
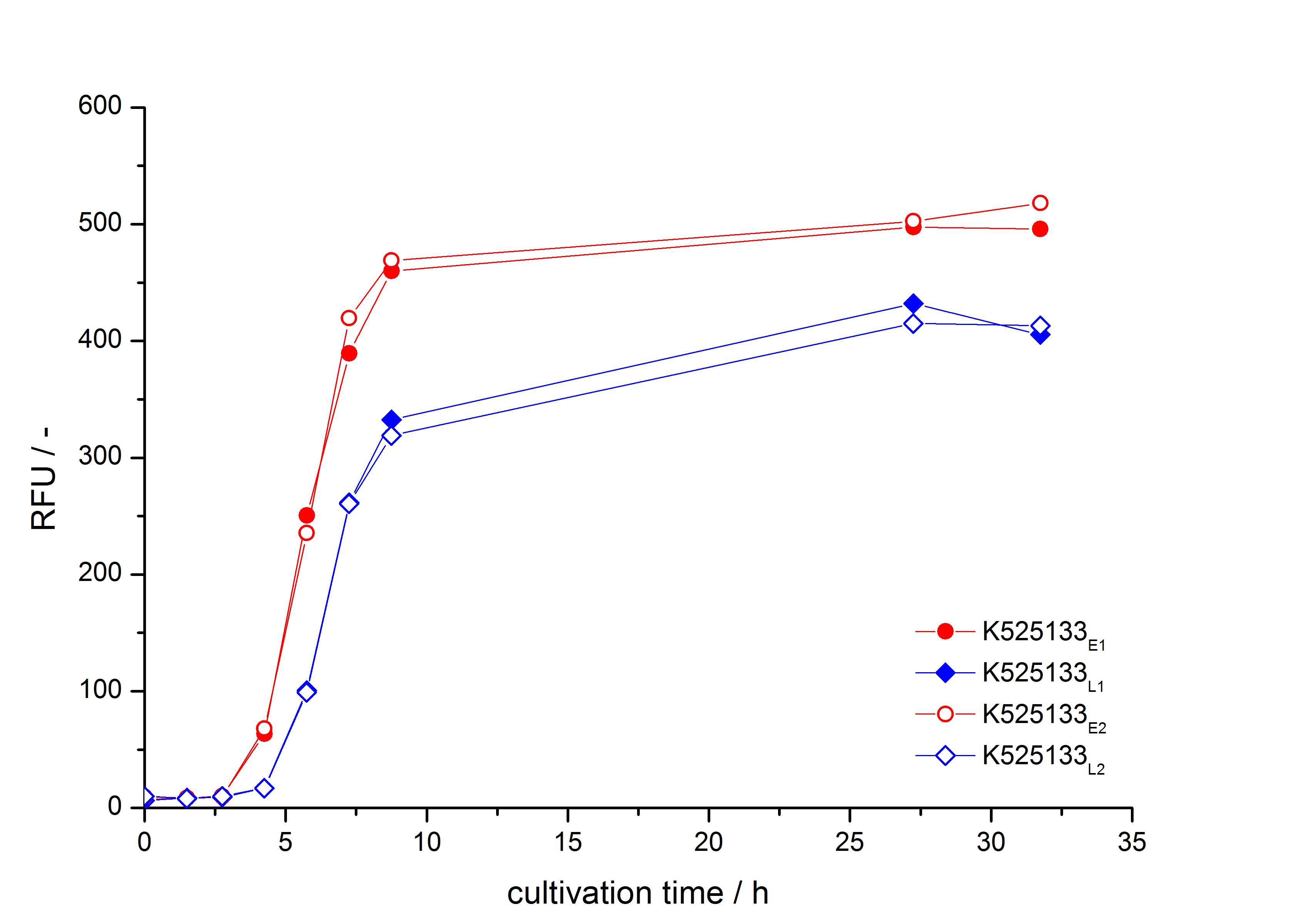
Bisphenol A:
- cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites
Week 8: 20th - 26th June
S-layer:
- successful cloning of K525232 using Gibson assembly.
- K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] without lipid anchor and TAT-sequence <partinfo>K525222</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010].
Organizational:
- finishing our first press release
- all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived
Week 9: 27th June - 3rd July
EXAMS !
Week 10: 4th July - 10th July
Bisphenol A:
- experiments on the influence of temperature, promoter strength and the characteristics of the fusion protein <partinfo>K123000</partinfo> || <partinfo>K123001</partinfo> on BPA degradation
S-layer:
- Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in E. coli KRX to test the functional efficiency of Corynebacterium TAT-sequence in E.coli and the effect of the autoinduction protocol. After one day of cultivation, an increased fluorescence in all periplasm fractions of E. coli expressing S-layer constructs with TAT-sequence could be recognized. This is an indication that the Corynebacterium TAT-signal sequence is functional in E. coli. Comparing the manual and the autoinduction protocol, no significant changes in fluorescence (after 14 h) are identifiable.
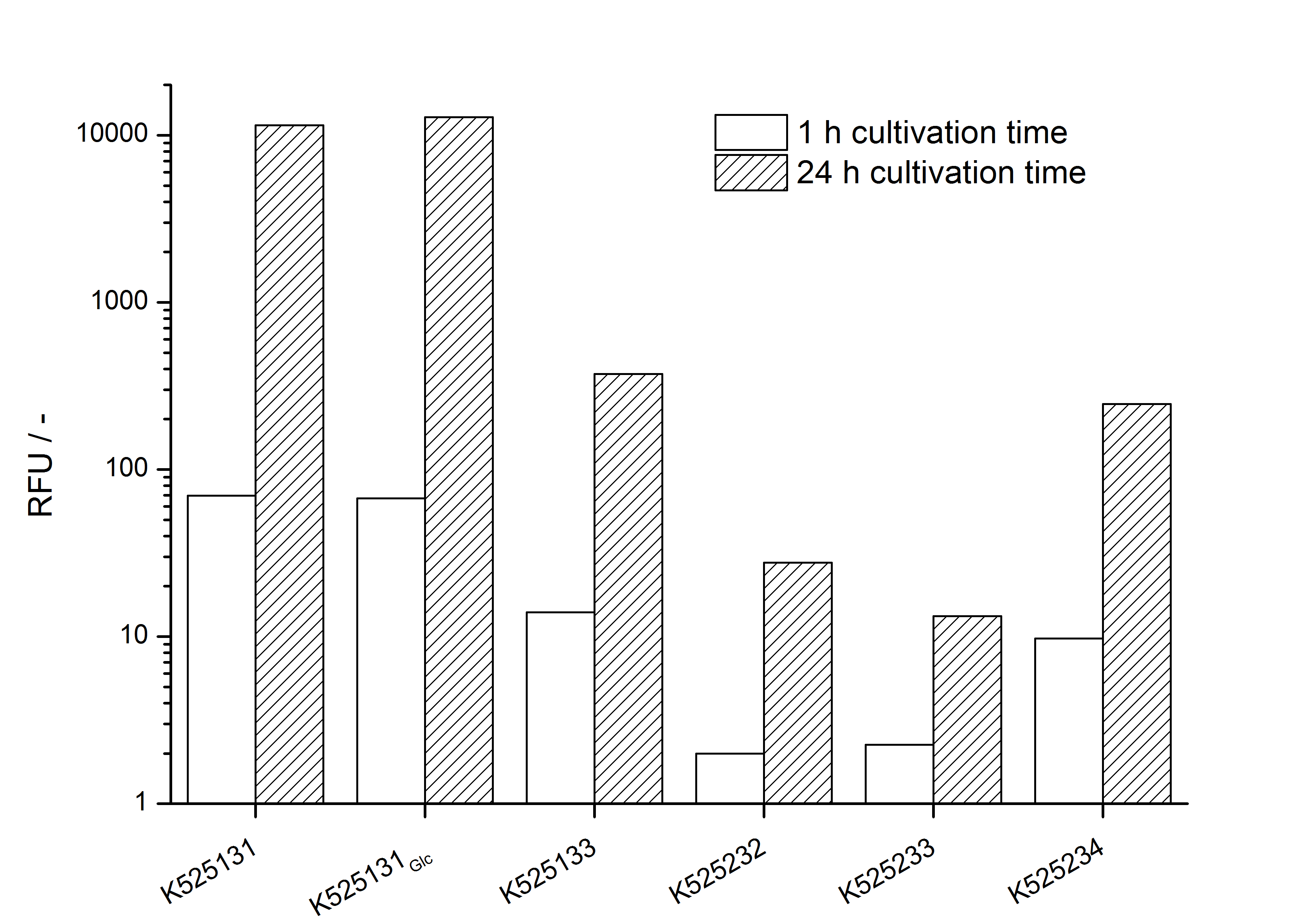

Organizational:
- presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research).
Week 11: 11th July - 17th July
Bisphenol A / S-layer:
- our BioBrick order (some fluorescent proteins and cleavage sites) from iGEM HQ arrived
S-layer:
- our synthesized S-layers SgsE and SbpA finally arrived
Organizational:
- presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos])
Week 12: 18th July - 24th July
Organizational:
- presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld
- meeting with the iGEM teams from Delft/NL, Edinburgh/UK, Odense/DK. Freiburg/DE and Ljubljana/SL at the Symposium ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627300144576/ Photos]) was fun!
Bisphenol A / S-Layer:
- removing illegal restriction sites to get valid BioBricks
NAD+ detection:
- cloning the NAD+-dependent DNA ligase from E. coli into BioBrick backbones
Week 13: 25th July - 31th July
S-layer:
- fusing the synthesized S-Layers to a bunch of fluorescent proteins
- successful cloning of K525231 using Gibson assembly.
- K525231: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] including lipid anchor and TAT-sequence <partinfo>K525221</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010].
- testing the basal transcription of E. coli KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD600 reveals the basal transcription of E. coli KRX. L-rhamnose in the medium leads to a ten times higher RFU per OD600.
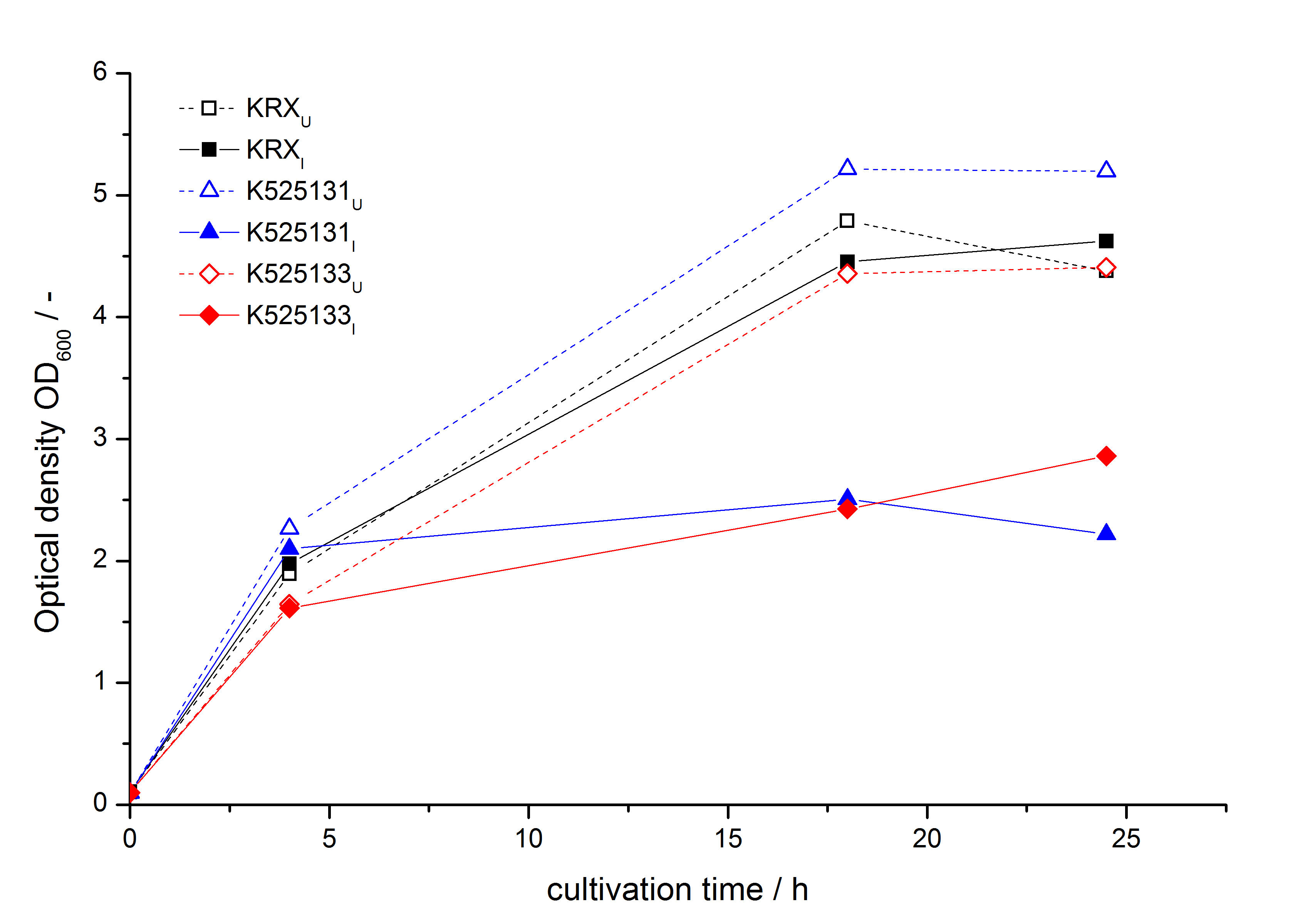

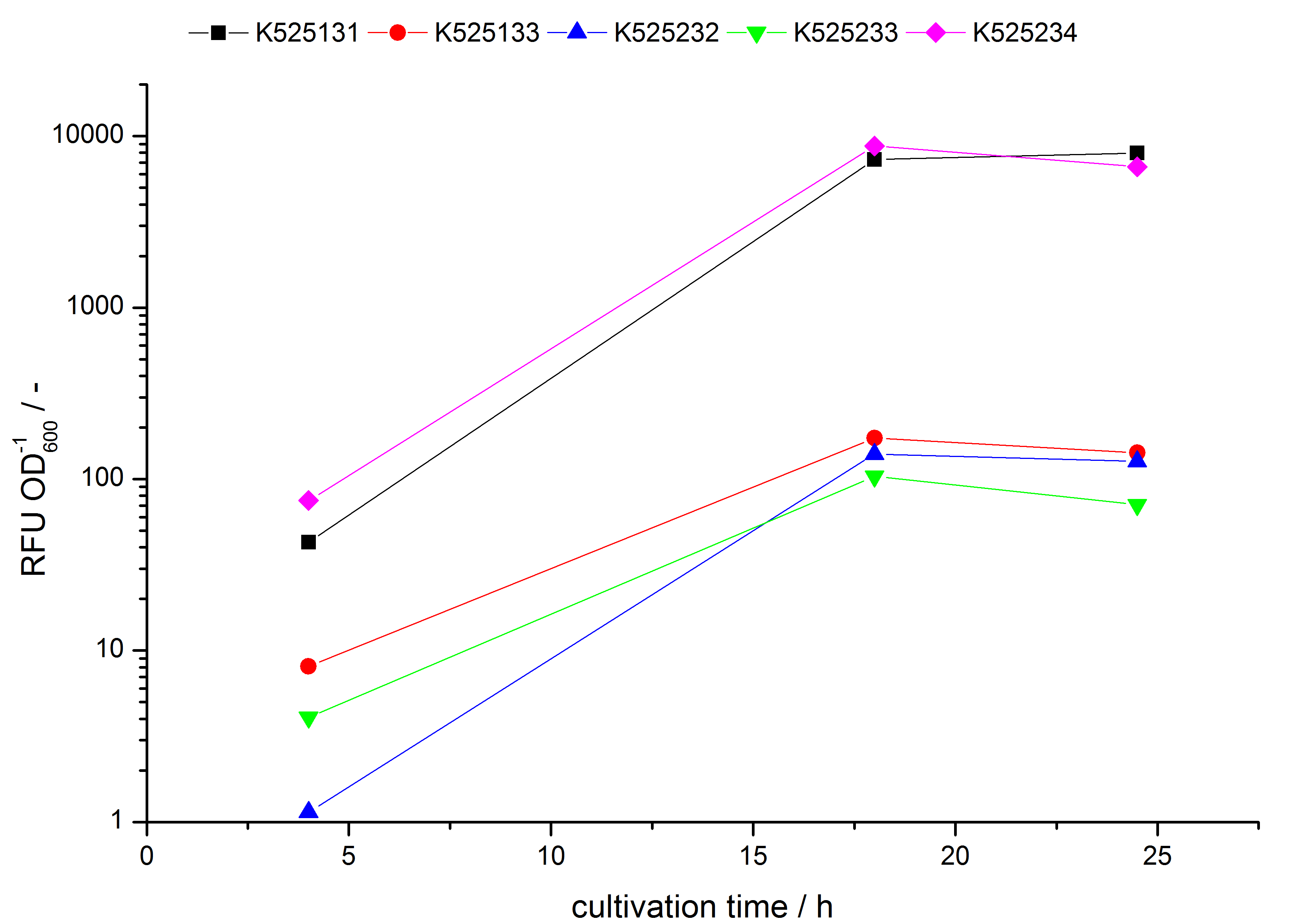
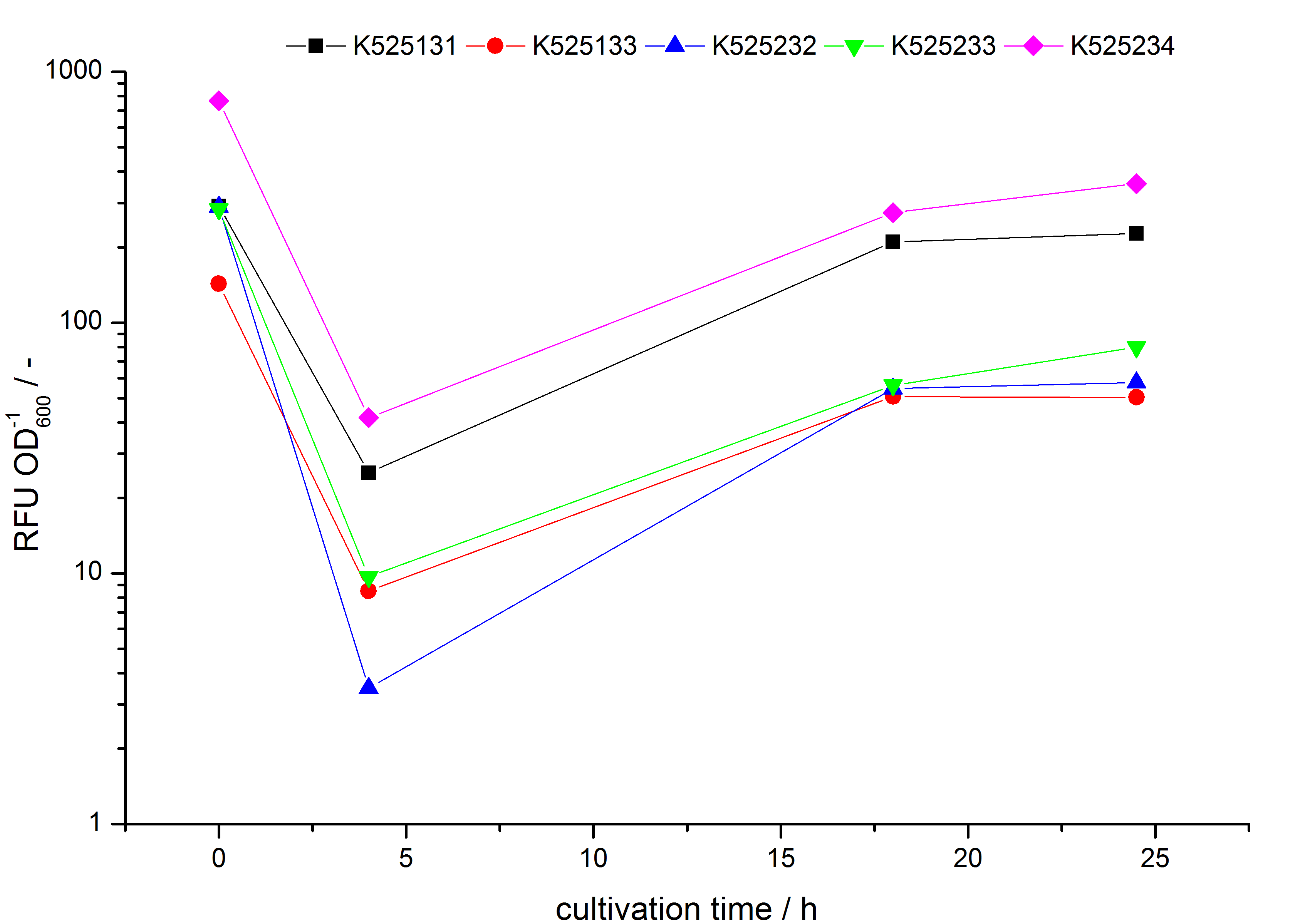
Bisphenol A:
- testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F)
Week 14: 1st August - 7th August
Bisphenol A:
- BPA analysis with extraction and LC-MS finally works and is very accurate
- Better results for BPA degradation in E. coli -> our fusion protein (<partinfo>K123000</partinfo> to <partinfo>K123001</partinfo>) can completely degrade BPA
- Measuring characterization results for different BPA degrading BioBricks
NAD+ detection:
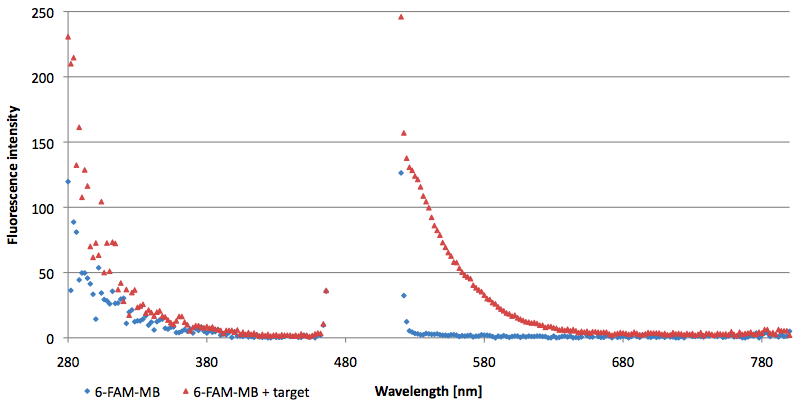
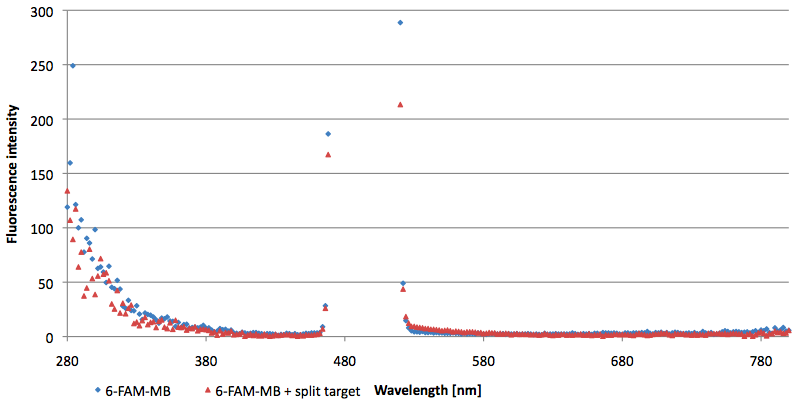
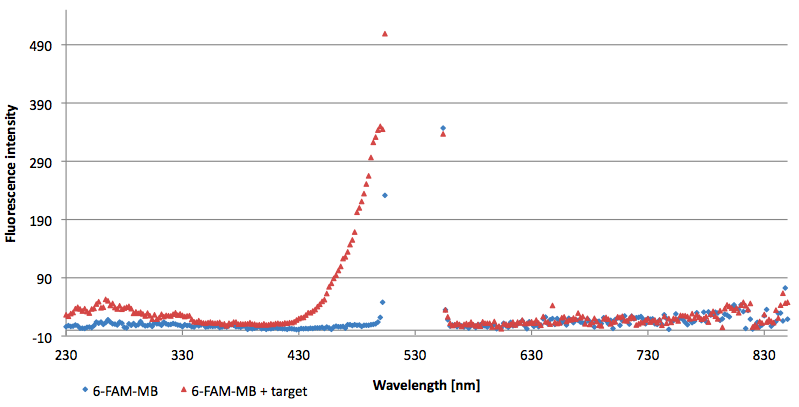
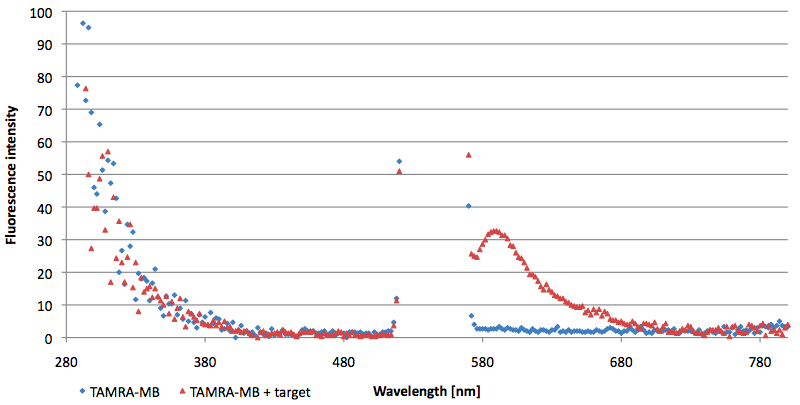
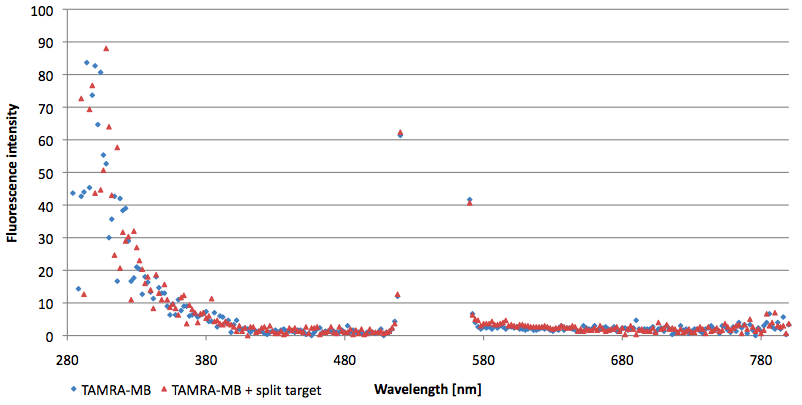
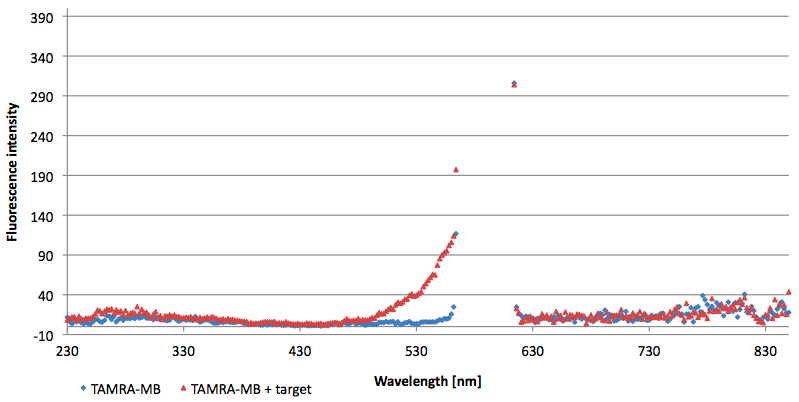
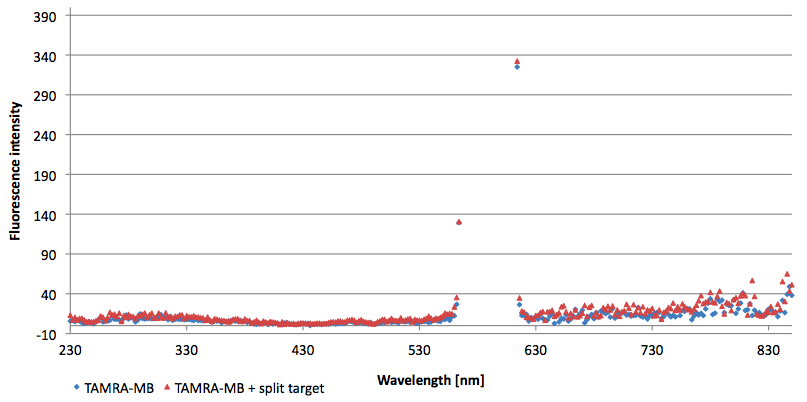
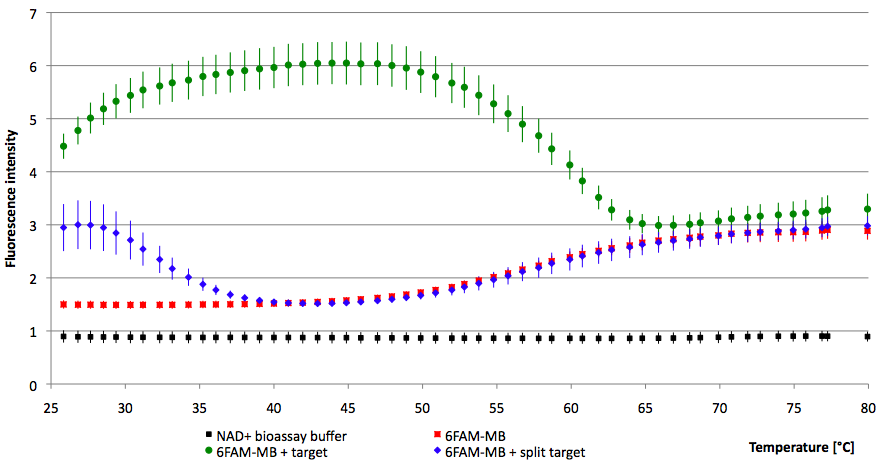
- Successful characterization of two differently labeled (6-FAM or TAMRA with Dabcyl) molecular beacons as a preparation for the NAD+ bioassay including autonomously produced NAD+-dependent DNA ligase (<partinfo>K525710</partinfo>) from E. coli (results shown below)
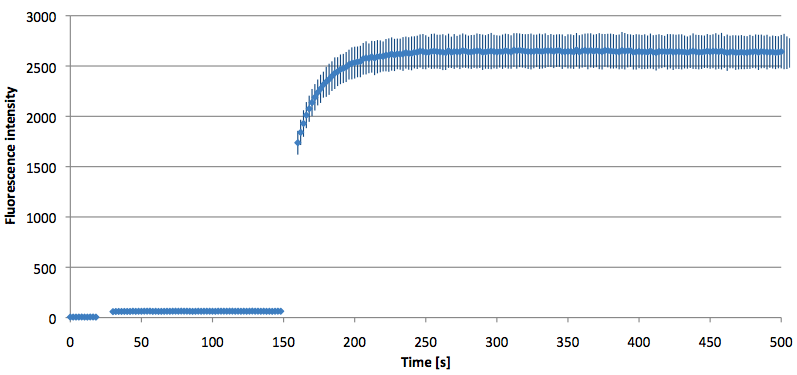
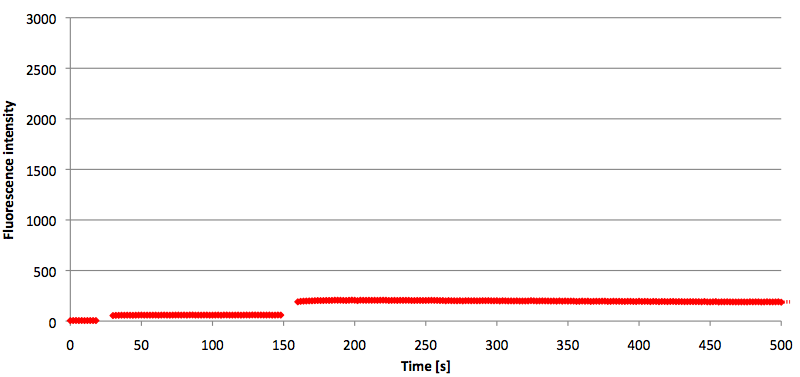
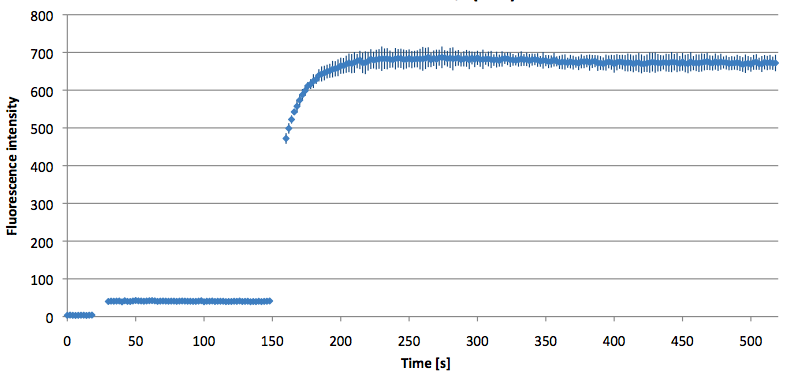
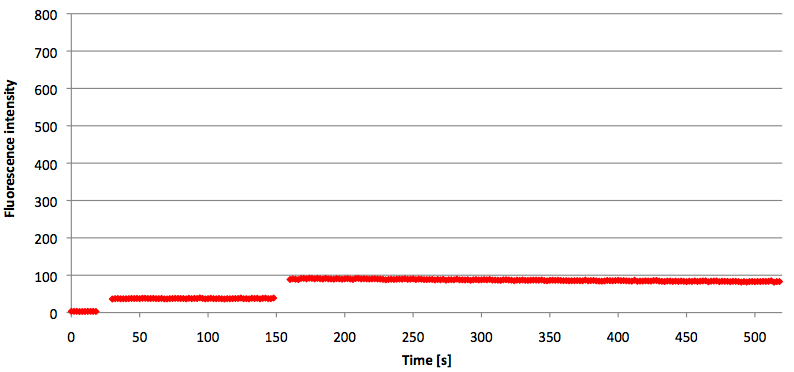
Week 15: 8th August - 14th August
Bisphenol A:
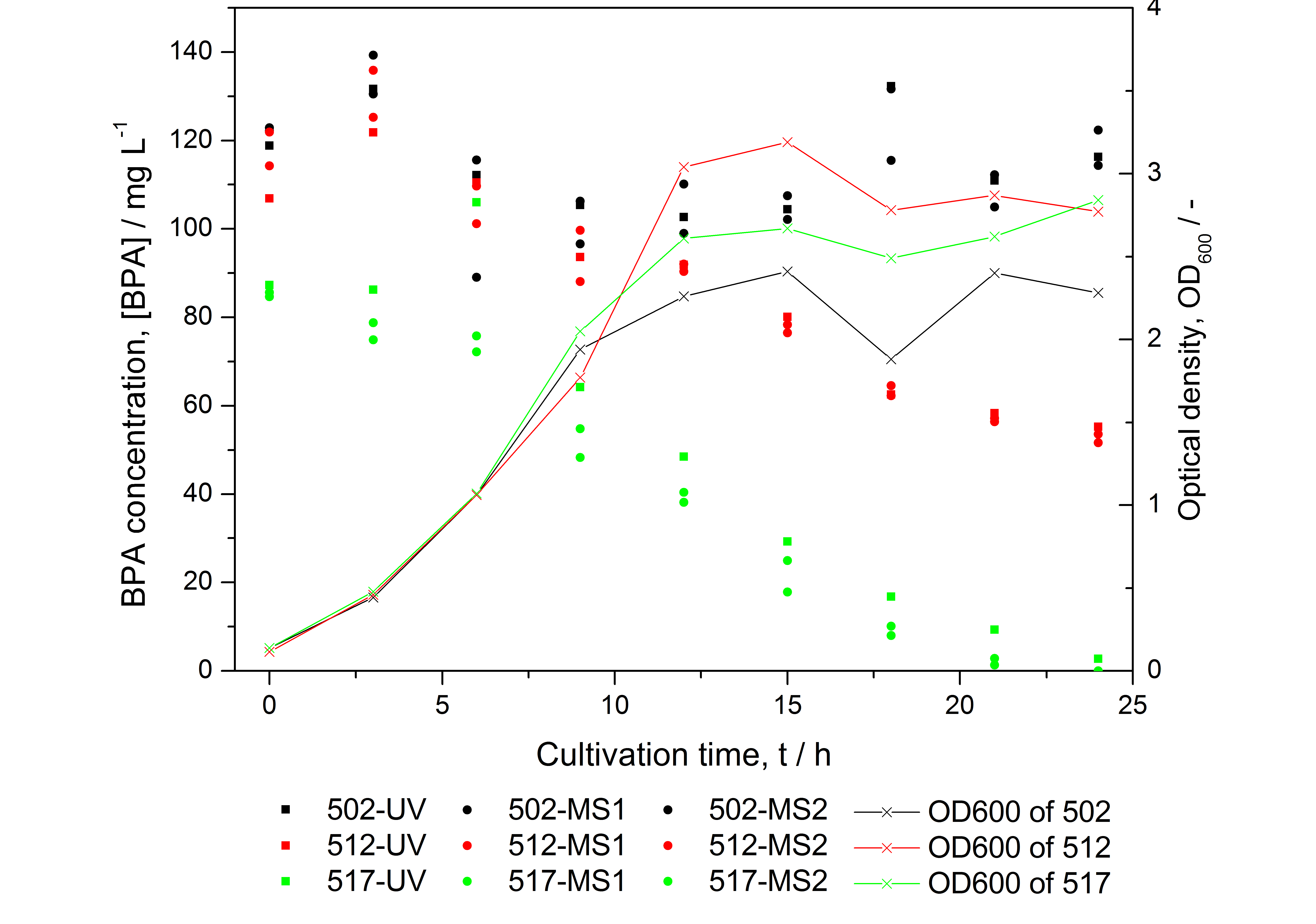
- BPA analysis with extraction and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare Figure 25)
- measuring of more samples from cultivations with BPA degrading BioBricks for further characterization
- we discovered some interesting results in our MS data - soon more
- testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins
- testing the influence of BPA on the growth of E. coli
- developing a model for BPA degradation by E. coli (compare Figure 26)
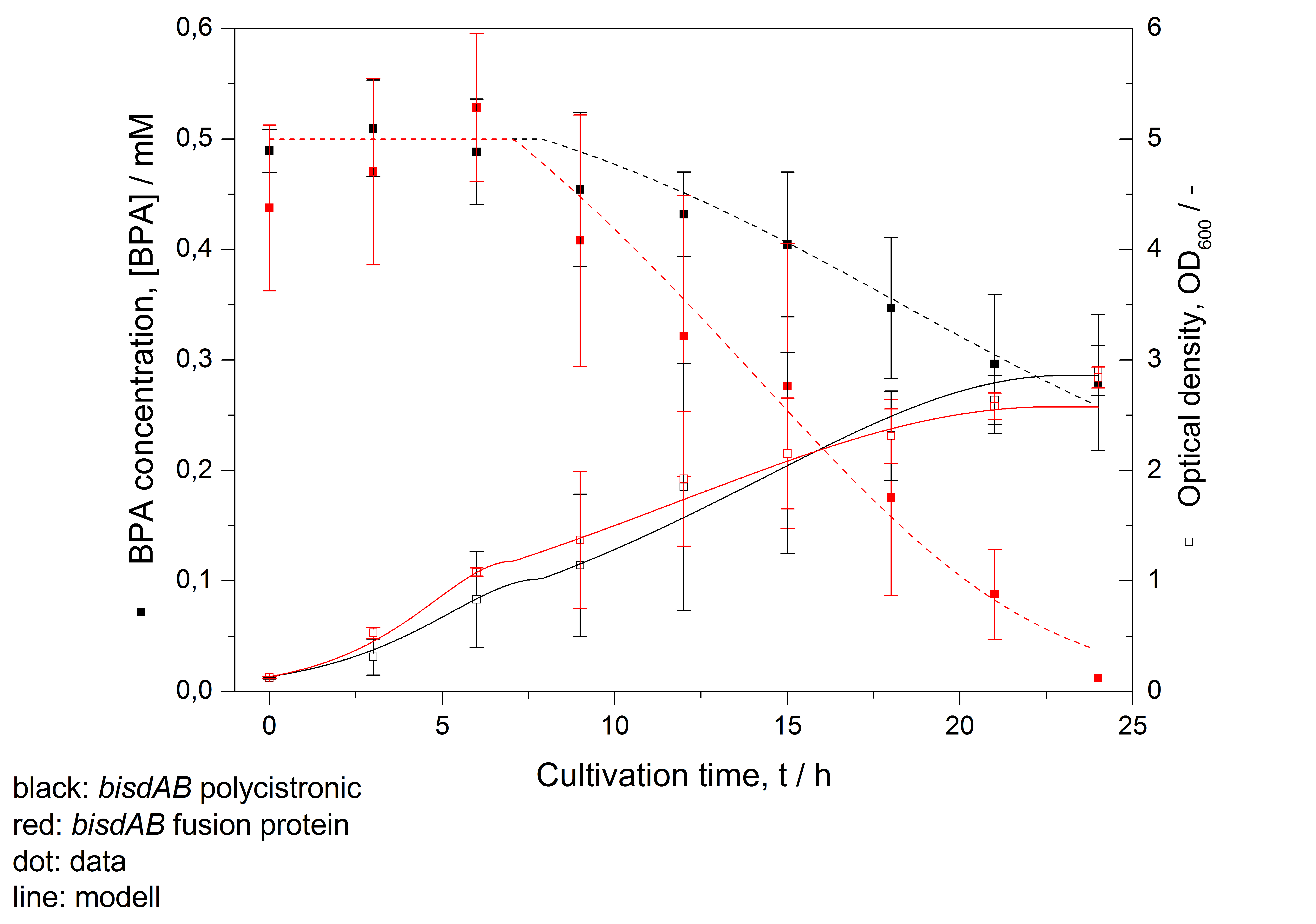
S-layer:
- MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by cspB gene) in E. coli KRX.
Week 16: 15th August - 21st August
S-layer:
- finally found our S-layer proteins in E. coli by analyzing the range of sizes in the polyacrylamid with a MALDI-TOF method-> now we can plan purification strategy
- fusion of FPs to sgsE successful
Bisphenol A:
- all metabolites of natural BPA degradation pathway found during degradation of BPA in E. coli by LC-MS -> now MS/MS to check structure of these metabolites to be sure
- some "metabolites" could also be found due to fragmentation during ionization of the product of BPA degradation
- testing whether E. coli can grow on BPA as the only carbon source (on M9 plates)
- seems like they can't
Week 17: 22nd August - 28th August
S-layer:
- cultivation and test purification of SgsE fusion proteins (<partinfo>K525304</partinfo>, <partinfo>K525305</partinfo> and <partinfo>K525306</partinfo>)
- SDS-PAGEs showed inclusion bodies formation in E coli KRX expressing sgsE.
Bisphenol A:
- purification of his-tagged BisdA and BisdB
- first characterization results entered into partsregistry
- cloning of fusion protein FNR:BisdA:BisdB successful
Week 18: 29th August - 4th September
S-layer:
- developing IEX clean-up for S-layers from Corynebacterium halotolerans
- Searching new purification methods for SgsE fusion proteins.
Bisphenol A:
- successful cloning of polycistronic FNR + BisdA + BisdB
NAD+ detection:
- Successful overexpression of NAD+-dependent DNA ligase (<partinfo>K525710</partinfo>) in E. coli KRX and purification with Ni-NTA columns utilizing the protein`s C-terminal His-tag. Trying out utility for the NAD+ bioassay now.
Organizational:
- participating at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld with a [http://www.geniale-bielefeld.de/programm/detail/date/456/show/Event/ lab tour] and a [http://www.geniale-bielefeld.de/programm/detail/date/295/show/Event/ presentation with public discussion afterwards (science café)]. Our participation is also mentioned in the regional newspaper [http://www.westfalen-blatt.de/nachricht/2011-08-30-wie-ein-hamster-im-laufrad/?cHash=b8421d55eb00ebf8532ebfa677a7e18f Westfalen Blatt] and in the internet on [http://www.direkt-bielefeld.de/Wissenschaft-im-Doppelpack-108856.html Bielefeld Direkt].
Week 19: 5th September - 11th September
S-layer:
- coating silica beads with fluorescent S-layer fusion protein SgsE | mCitrine, remove these proteins again and find them in SDS-PAGE
- characterizing immobilization behaviour of S-layers on silica beads -> seems to work (Figure 27)
- cultivate bigger amounts of and develop new purification strategy for S-layer fusion proteins
- testing purification of S-layer fusion proteins with his-tag
- cultivations of fusion protein between NADP+-ferredoxin oxidoreductase and SgsE / SbpA
- also SbpA | mCitrine fusion protein
NAD+ detection:
- Suitability of autonomously produced DNA ligase (<partinfo>K525710</partinfo>) from E. coli for the NAD+ bioassay could be demonstrated after treatment with NMN for deadenylation. Checking out the limit of detection right now.
Week 20: 12th September - 18th September
S-layer:
- testing a purification and coating protocol for S-layer fusion protein mCitrine|SbpA.
- characterizing immobilization behaviour of mCitrine|SbpA [http://partsregistry.org/Part:BBa_K525405 (K525405)] on silica beads
- optimizing purification and coating protocol for SgsE fusion proteins.
- last characterization cultivations of [http://partsregistry.org/Part:BBa_K525304 K525304], [http://partsregistry.org/Part:BBa_K525305 K525305], [http://partsregistry.org/Part:BBa_K525306 K525306], [http://partsregistry.org/Part:BBa_K525405 K525405] and [http://partsregistry.org/Part:BBa_K525406 K525406].
Bisphenol A:
- successful cloning of fusion protein FNR + BisdA | BisdB and polycistronic FNR + BisdA + BisdB behind constitutive promoter -> characterizing these last constructs
NAD+ detection:
- NAD+ assay works :)
Week 21: 19th September - 25th September
Organizational:
- lab work is finished, the last measurements and cloning experiments into pSB1C3 are done
- now it is time to fill the Wiki and Partsregistry with brilliant data until European Wikifreeze @ September 21st 11:59 pm EDT!
Week 22: 26th September - 2nd October
Preparation of the presentation and the poster for the European Regional Jamboree.
- practicing the presentation with the Coryne and fermentation engineering working group as audience.
We are qualified for the final in Boston!!! Now labwork can continue.
Week 23: 3rd October - 9th October
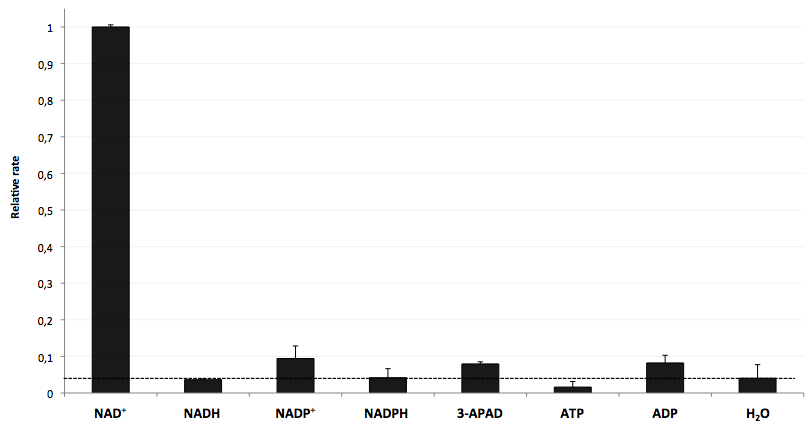
S-layer:
- scale-up cultivation of E. coli expressing [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] fusion protein.
- inclusion body purification and testing further purification strategies.
- assembly of [http://partsregistry.org/Part:BBa_K525422 K525422] containing mCitrine|SbpA fused with a His-tag ([http://partsregistry.org/Part:BBa_K157011 K157011]) for easer purification.
- assembly of [http://partsregistry.org/Part:BBa_K525311 K525311] consisting of the SbpA S-layer fused with firefly-luciferase ([http://partsregistry.org/Part:BBa_K525999 K525999]) for testing an enzyme reaction on immobilized S-layer proteins.
- ordering silicon wafer as a new surface for immobilization.
- successful cloning of fusion protein of K525303 and K525560 [http://partsregistry.org/Part:BBa_K525001 (K525001)],[http://partsregistry.org/Part:BBa_K525311 (K525311)], [http://partsregistry.org/Part:BBa_K525006 SbpA|LigA (K525006)], [http://partsregistry.org/Part:BBa_K525422 (K525422)],[http://partsregistry.org/Part:BBa_K525322 (K525322)]
NAD+ detection:
- testing selectivity of LigA (<partinfo>BBa_K525710</partinfo>) for its substrate NAD+ to verify further applications of the molecular beacon based NAD+ bioassay dealing with NAD+ detection in analyte mixtures such as cell lysates.
Week 24: 10th October - 16th October
S-layer:
- first expression of K525422 in E. coli KRX and His-tag affinity chromatography based purification using denaturing conditions.
- Test purifications were carried out using binding buffer with and without imidazole.
- new inclusion body purification of K525305 and K525405 to get new biological material to test the further purification methods.
- establishment of ion exchange chromatography (IEX) and hydrophobic interaction chromatography (HIC) as futher purification steps for S-layer proteins from Lysinibacillus sphaericus and Geobacillus stearothermophilus.
- diethylaminoethyl cellulose (DEAE) as IEX and butyl sepharose™ 4 Fast Flow as HIC purification media.
- test and scale-up expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] in E. coli KRX.
- after enzymatic cell lysis with lysozyme and inclusion body purification highest luminescence was measured in the lysis supernatant. This implicates that the fusion protein does not form inclusion bodies.
- successful cloning of fusion protein of CspB and luciferase with Gibson assembly
NAD+ detection:
- the NAD+ bioassay was successfully coupled to the NADH-dependent reaction of lactic acid dehydrogenase and could therefore be used to quantify pyruvate
BPA-degradation:
- successful cloning of [http://partsregistry.org/Part:BBa_K525562 (K525562)] ( Fusion protein of [http://partsregistry.org/Part:BBa_K525499 (K525499)], [http://partsregistry.org/Part:BBa_K123000 (K123000)]and [http://partsregistry.org/Part:BBa_K123001 (K5123001)] with middle strong promoter) and [http://partsregistry.org/Part:BBa_K525534 (K525534)]
Week 25: 17th October - 23th October
S-layer:
- new expression of <partinfo>K525311</partinfo> to develop an individual purification strategy without inclusion body purification.
- testing a new purification strategy for the fusion protein of the SgsE and the firefly-luciferase based on the IEX and HIC
- first expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] and successful His-tag affinity chromatography based purification using denaturing conditions.
NAD+ detection:
- testing LigA for molecular cloning
Week 26: 24th October - 30th October
S-layer:
- test expression of [http://partsregistry.org/Part:BBa_K525534 reductase|His-tag (K525534)] in E. coli KRX. and testing non denaturating His-tag affinity chromatography.
- recrystallization of [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] on silicon wafer using two different immobilization strategies.
- trying to get pictures of the S-layer nanostructures on coated silicon wafers using reflection electron microscope (REM) and atomic force microscopy (AFM)
- not enough time left to find a successful method
 "
"