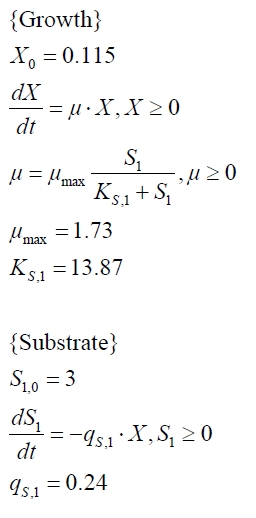Team:Bielefeld-Germany/SinceAmsterdam
From 2011.igem.org


Contents |
What we improved since the European Regional Jamboree
BPA degradation
Specificity of bisphenol A degradation with E. coli
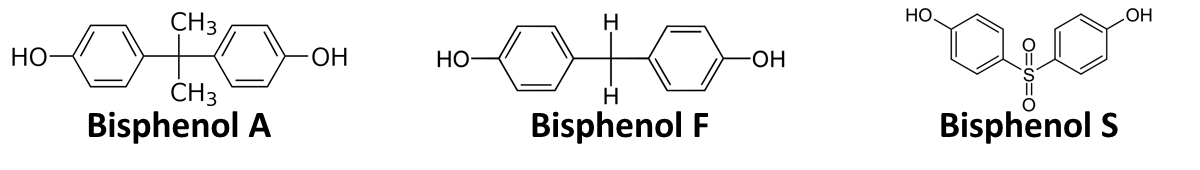
In order to access the specificity of the bisphenol A degradation by the bisdA | bisdB fusion protein, we tested how well two similar bisphenols, bisphenol F (BPF) and bisphenol S (BPS), were utilized. The structure of those bisphenols differs only in the chemical group linking the two phenols from that of bisphenol A (see Figure 1).
BPF and BPS are used in a broad range of applications that involve the use of polycarbonates or epoxy resins and thus can often be found were BPA is also present. Accordingly their presence is a potential disruptive factor that could lead to a false positive signal with our biosensor. This is especially true for BPS that in some cases is used as a substitute for BPA in baby bottles [http://www.nytimes.com/2011/04/18/business/global/18iht-rbog-plastic-18.html]. Studies concerning the environmental pollution with BPF ([http://www.sciencedirect.com/science/article/pii/S0043135401003670 Fromme et al. (2002)]) and the acute toxicity, mutagenicity and estrogenicity of BPF and BPS ([http://onlinelibrary.wiley.com/doi/10.1002/tox.10035/abstract Chen et al. (2001)] and [http://toxsci.oxfordjournals.org/content/84/2/249 Kitamura et al. (2005)]) indicate their potential harmfulness but further research is needed to fully assess their impact on human health.
E. coli KRX carrying genes for the bisdA | bisdB fusion protein behind the medium strong constitutive promoter <partinfo>J23110</partinfo> with RBS <partinfo>B0034</partinfo> was cultivated at 30 °C for 36 h with LB-Medium containing 120 mg L-1 BPA, BPF respectively BPS. The BPF and BPS concentration were determined by HPLC using the same method as with BPA. Figure 2 shows the degradation of the respective bisphenol after 24 h of cultivation in percent.
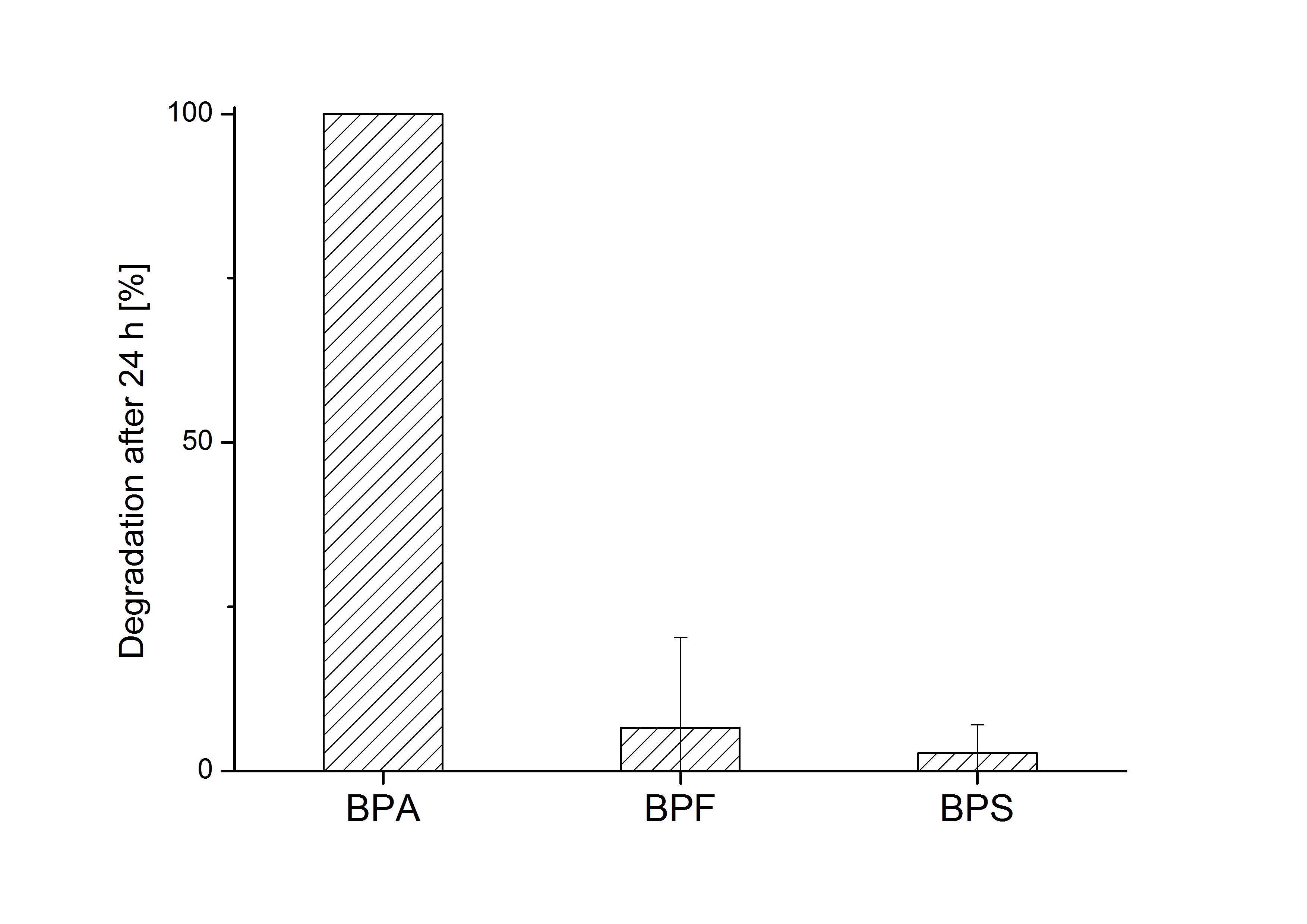
The results of the experiment indicate that the bisdA | bisdB fusion protein has a high specificity for the degradation of BPA. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of E. coli KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely.
BPA degradation with constructs containing FNR
Constructs containing FNR, BisdA and BisdB polycistronic (<partinfo>K525551</partinfo>), FNR and a fusion protein of BisdA and BisdB (<partinfo>K525582</partinfo>) and a fusion protein of FNR, BisdA and BisdB (<partinfo>K525560</partinfo>) were tested for their ability to degrade BPA. Cultivations were done using the same conditions as with previous constructs. The results are shown in Figure 3 below.
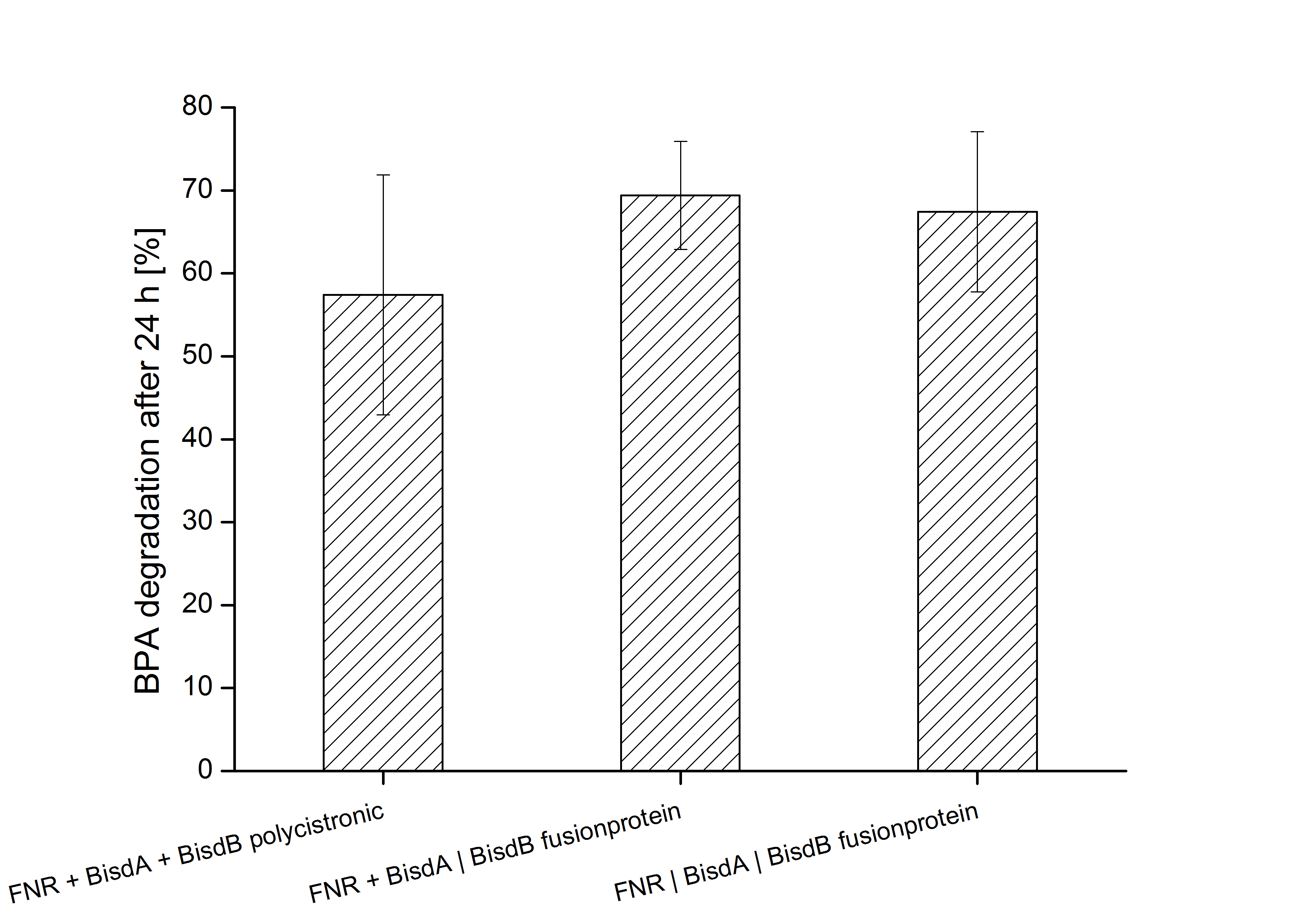
The results indicate that the BPA degradation was improved using fusion proteins compared to the completely polycistronic construct. Also the fusion protein of all three enzymes (<partinfo>K525560</partinfo>) involved in the degradation of BPA worked as intended which is quite impressive.
Comparison of all constructs used for BPA degradation
Figure 4 gives an overview of the constructs employed for the degradation of BPA.
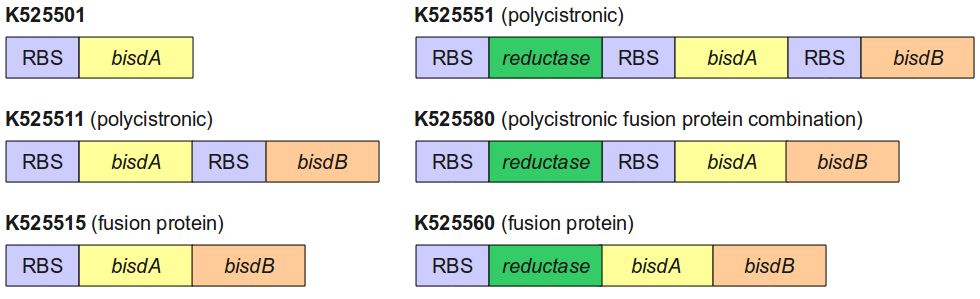
All constructs were cultivated under the same conditions as described under Cultivations. BPA concentrations were measured using HPLC. Figure 5 shows the percentage of BPA degraded in 24 h of cultivation and the specific BPA degradation rate calculated with the regular model and in the case of <partinfo>K525560</partinfo> with the model for the fusion protein between FNR, BisdA and BisdB.

We are especially impressed that the fusion protein of FNR, BisdA and BisdB (<partinfo>K525560</partinfo>) not only was capable to degrade BPA but also was the fastest construct we employed. This also contributes to the feasibility of our approach for a cell free BPA biosensor since in a cell free environment fusion proteins are beneficial when compared to their non-fused counter parts.
Model for the fusion protein between FNR, BisdA and BisdB
The cultivations and BPA degradation of E. coli KRX carrying a fusion protein consisting of ferredoxin-NADP+ oxidoreductase, ferredoxinbisd and cytochrome P450bisd differ from the cultivations with the other BPA degrading BioBricks. Thus, the model for these cultivations has to be adjusted to this behaviour. First of all, no diauxic growth is observed so the growth can be modeled more easily like
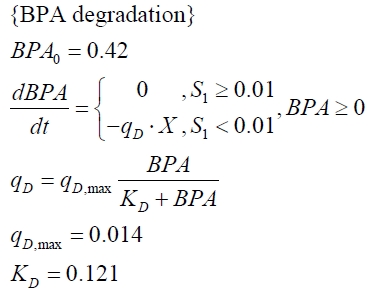
The BPA degradation starts when the imaginary substrate is depleted, like observed in the other cultivations with BPA degrading BioBricks. But it seems that the BPA degradation is getting slower with longer cultivation time. So it is modeled with a Monod-like term in which the specific BPA degradation rate is dependent on the BPA concentration:
with the maximal specific BPA degradation rate qD,max and the constant KD.
Figures 6-10 show a comparison between modeled and measured data for cultivations with BPA degrading fusion protein E. coli. In Table 1 the parameters for the model are given, obtained by curve fitting the model to the data.
Comparison between modeled and measured data
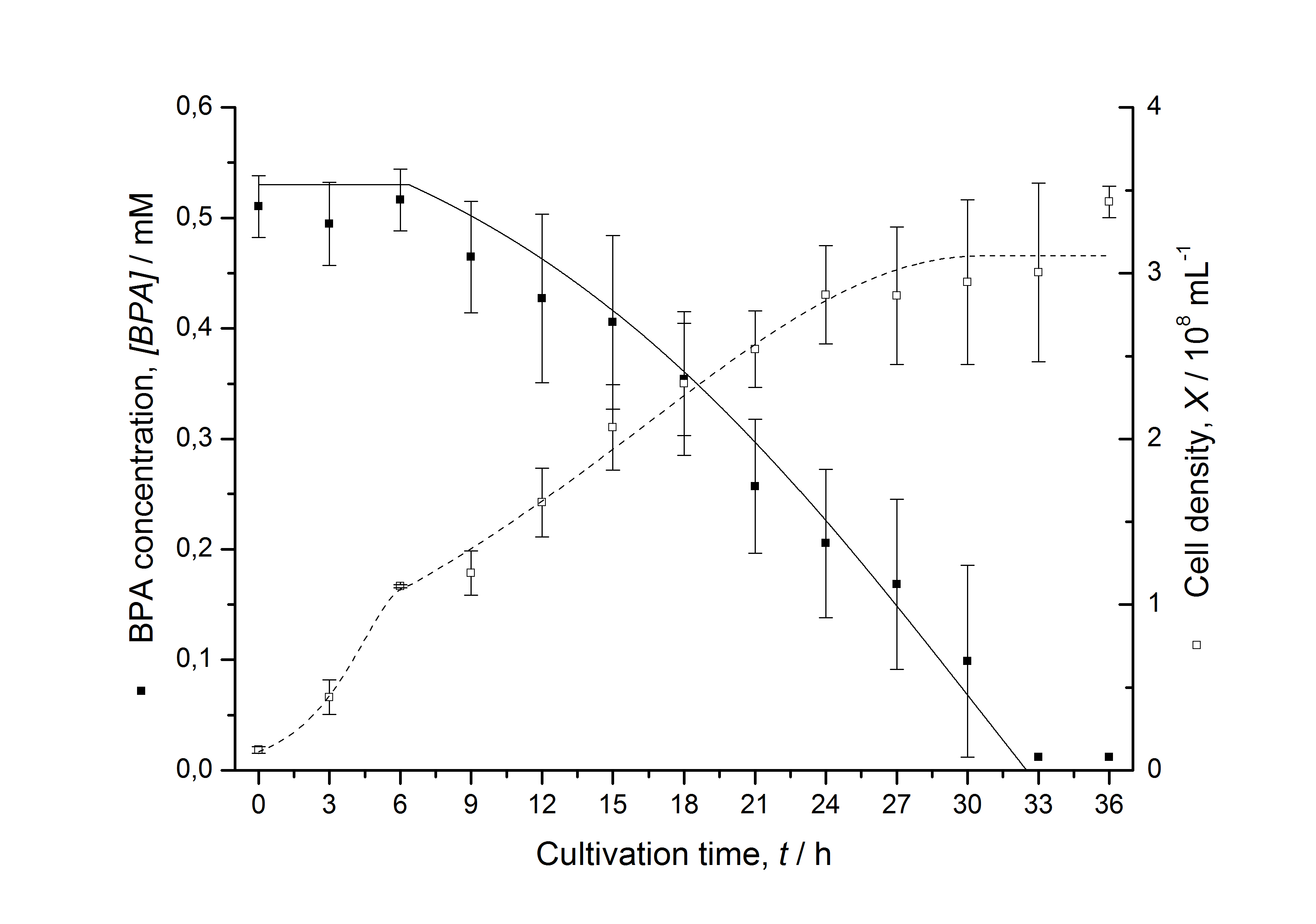
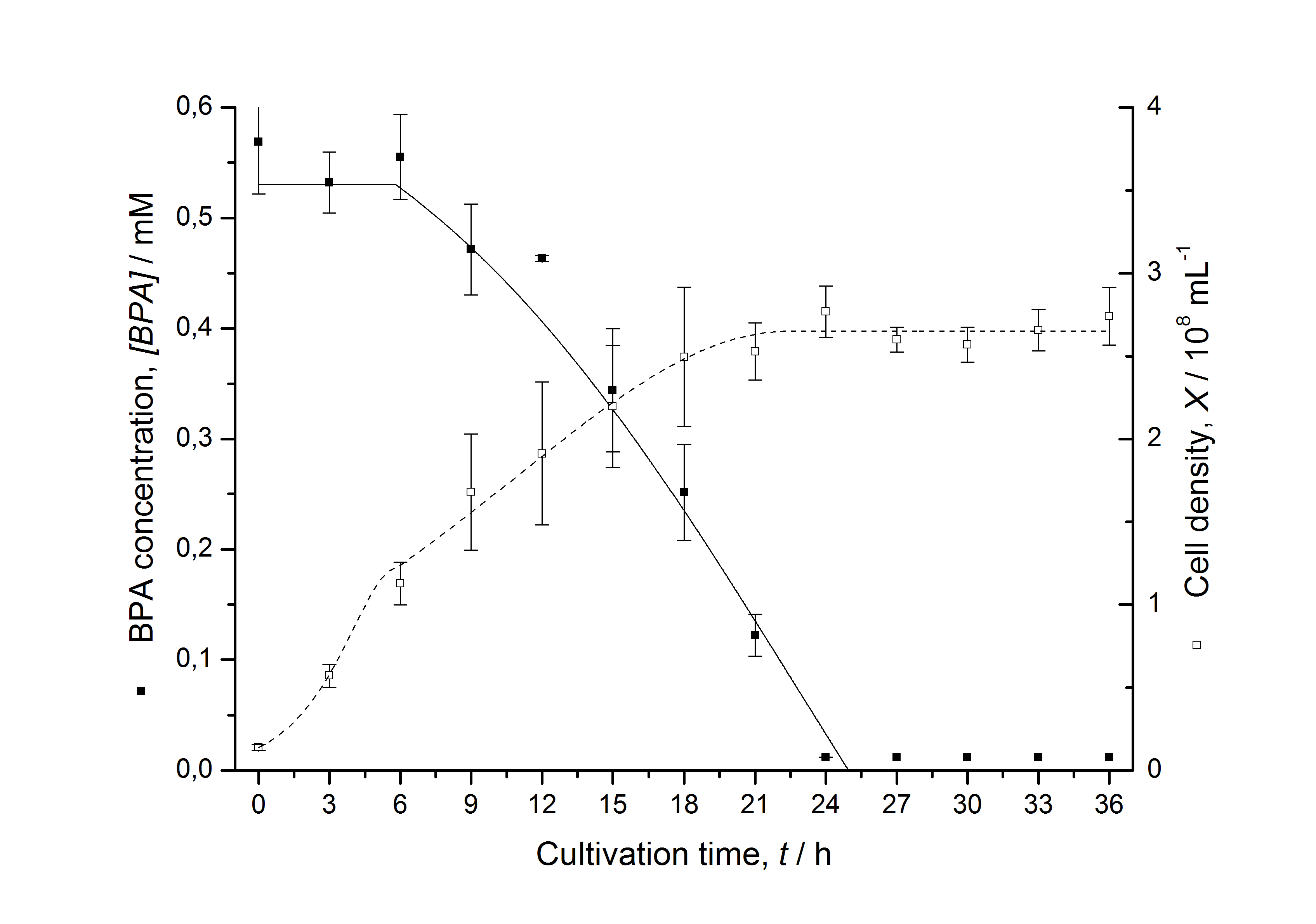
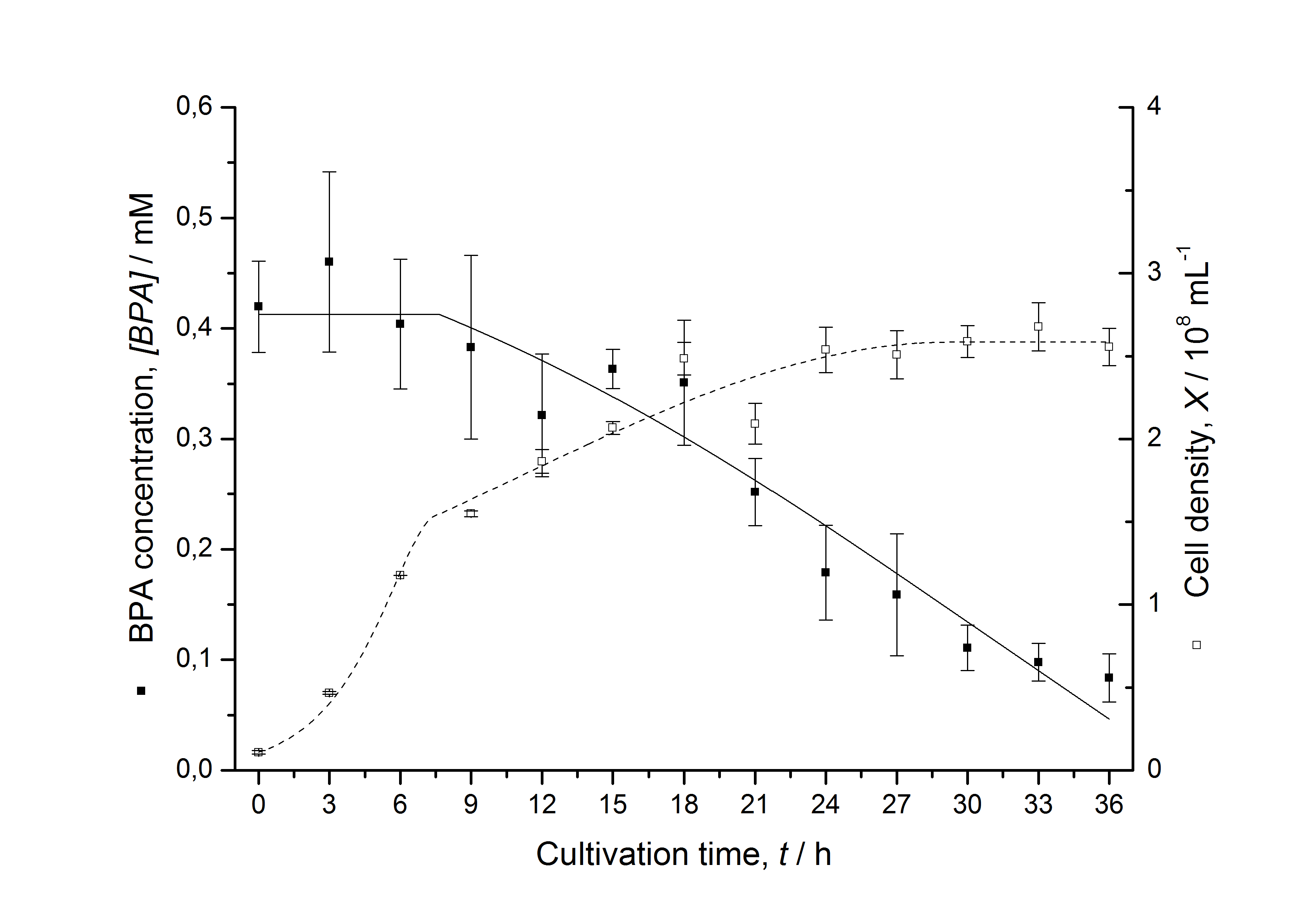
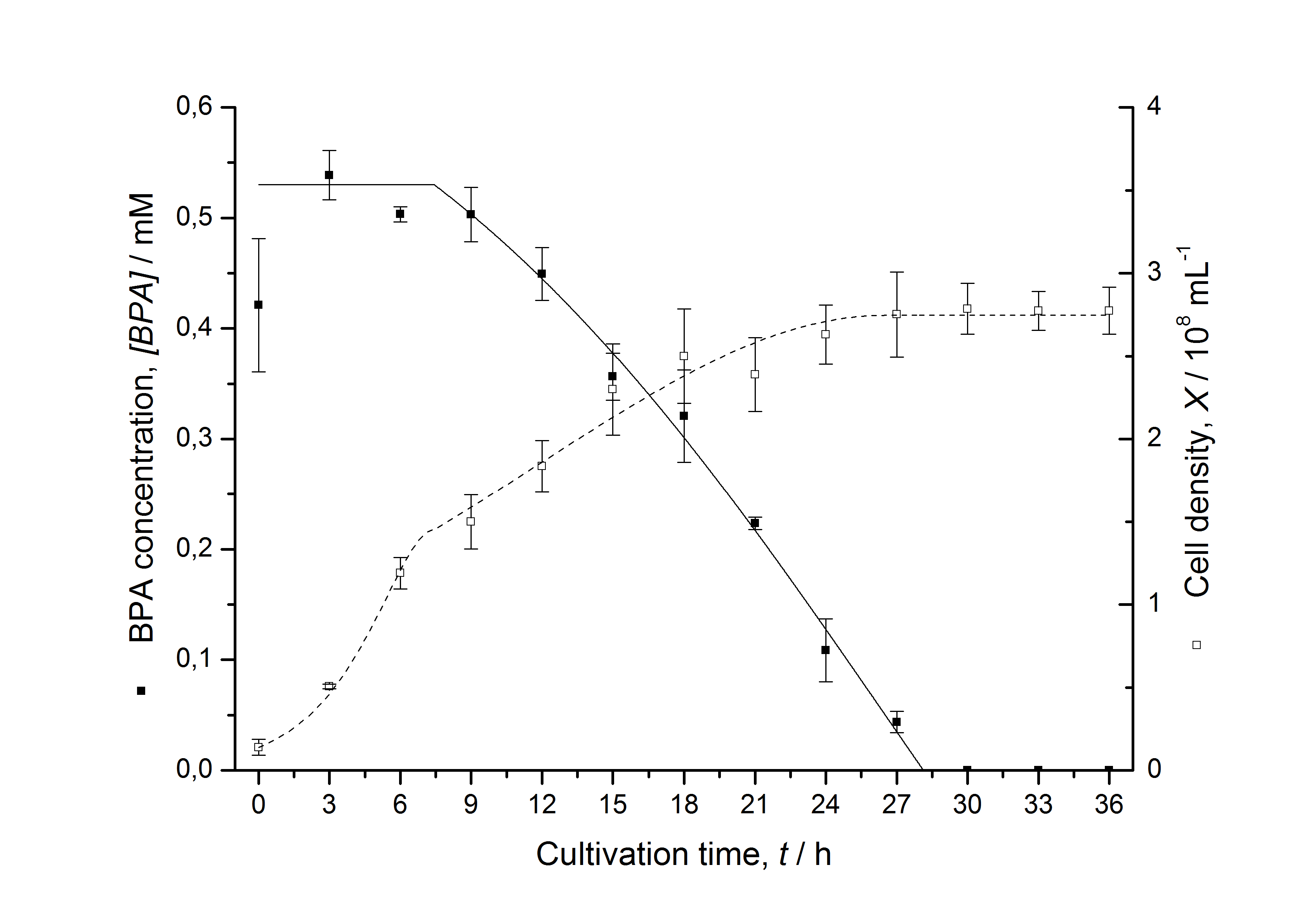
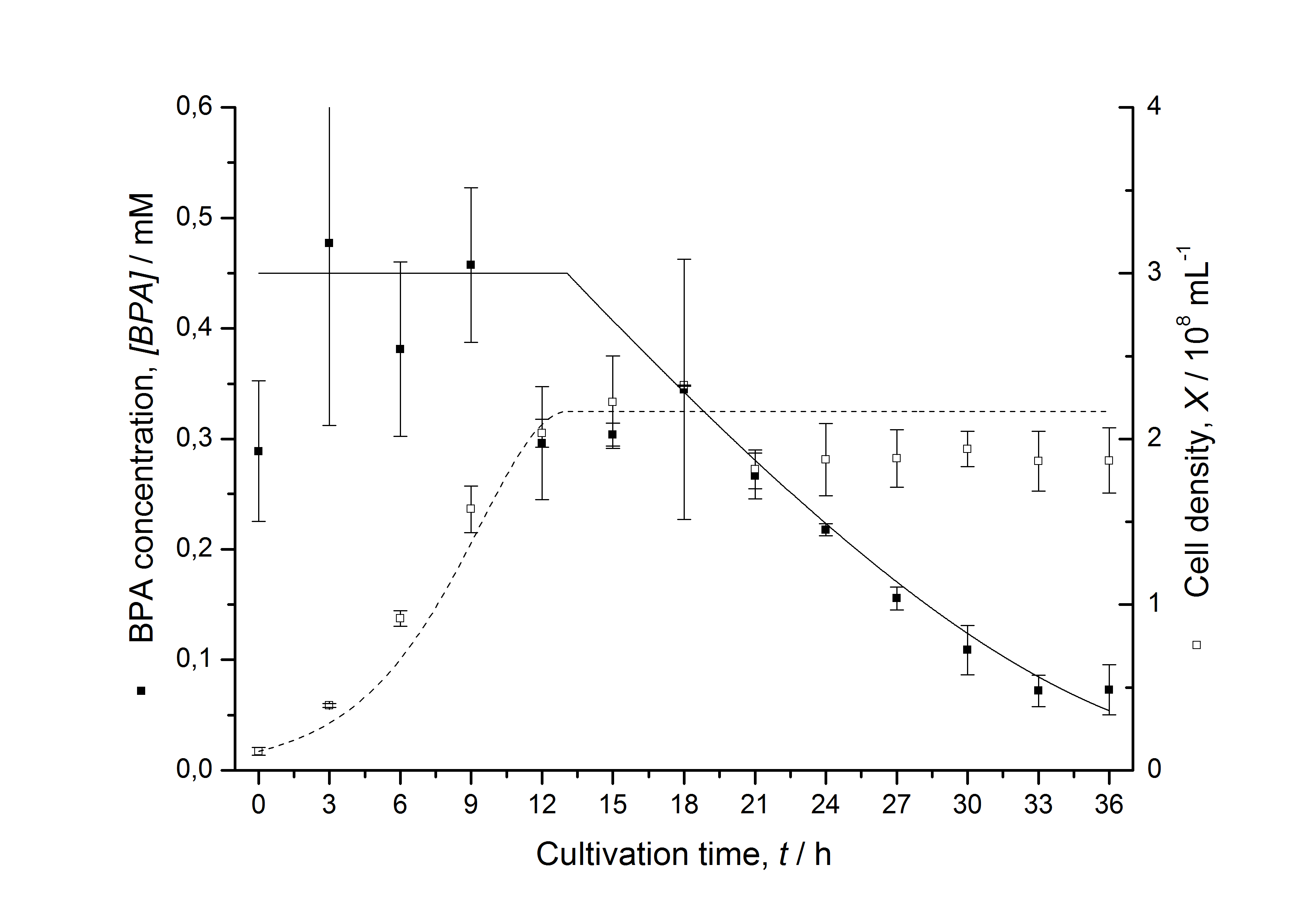
Table 1: Parameters of the model.
| Parameter | <partinfo>K525512</partinfo> | <partinfo>K525517</partinfo> | <partinfo>K525552</partinfo> | <partinfo>K525562</partinfo> | <partinfo>K525582</partinfo> |
|---|---|---|---|---|---|
| X0 | 0.112 108 mL-1 | 0.138 108 mL-1 | 0.109 108 mL-1 | 0.115 108 mL-1 | 0.139 108 mL-1 |
| µmax | 1.253 h-1 | 1.357 h-1 | 0.963 h-1 | 1.730 h-1 | 0.858 h-1 |
| KS,1 | 2.646 AU-1 | 1.92 AU-1 | 5.35 AU-1 | 13.87 AU-1 | 3.05 AU-1 |
| KS,2 | 265.1 AU-1 | 103.1 AU-1 | 82.6 AU-1 | - | 32.5 AU-1 |
| S1,0 | 1.688 AU | 1.166 AU | 4.679 AU | 3.003 AU | 2.838 AU |
| qS,1 | 0.478 AU 10-8 cell-1 | 0.319 AU 10-8 cell-1 | 0.883 AU 10-8 cell-1 | 0.240 AU 10-8 cell-1 | 0.544 AU 10-8 cell-1 |
| S2,0 | 16.091 AU | 6.574 AU | 3.873 AU | - | 2.402 AU |
| qS,2 | 0.295 AU 10-8 cell-1 | 0.191 AU 10-8 cell-1 | 0.082 AU 10-8 cell-1 | - | 0.056 AU 10-8 cell-1 |
| BPA0 | 0.53 mM | 0.53 mM | 0.41 mM | 0.45 mM | 0.53 mM |
| qD | 8.76 10-11 mM cell-1 | 1.29 10-10 mM cell-1 | 5.67 10-11 mM cell-1 | - | 1.13 10-10 mM cell-1 |
| qD,max | - | - | - | 1.32 10-10 mM cell-1 | - |
| KD | - | - | - | 0.121 mM-1 | - |
The specific BPA degradation rate per cell qD is about 50 % higher when using the fusion protein compared to the polycistronic bisdAB gene. On average, this results in a 9 hours faster, complete BPA degradation by E. coli carrying <partinfo>K525517</partinfo> compared to <partinfo>K525512</partinfo> as observed during our cultivations. The fusion protein between BisdA and BisdB improves the BPA degradation by E. coli. Introducing a polycistronic ferredoxin-NADP+ reductase gene into these systems does not lead to a higher specific BPA degradation rate. The rates are a bit lower, though. The effect that the BisdA | BisdB fusion protein is more efficient than BisdB when expressed polycistronically with BisdA can still be observed in this setup. The cultivations with the expression of the fusion protein FNR | BisdA | BisdB differ from the expression of the other BPA degrading BioBricks. The BPA degradation rate is concentration dependent which is typical for enzymatic reactions but was not observed in the other cultivations. In addition, the growth was faster and not diauxic. The maximal specific BPA degradation rate of this BioBrick is higher than the observed specific BPA degradation rates in the other cultivations. But due to the dependence of the BPA degradation from the BPA concentration the BPA degradation with this BioBrick is not as efficient and not complete. Anyway, the fact that this BioBrick is working, is impressive.
S-layer
Purification
The purification strategy could be accelerated by using a His-6-affinity tag and is more simple and pure now.
Scheme of purification strategy for SgsE (fusion) proteins with His-tag:
By fusing the SgsE | mCitrine with a C-terminal [http://partsregistry.org/Part:BBa_K157011 His-6-tag] the S-layer protein could be simply purified by using a denaturating His-tag affinity chromatography. This purification strategy has the advantage that no time-consuming and complex inclusion body purification and filtration is necessary to decrease the amount of native E. coli proteins. Additional to the simplification, a higher purity of the S-layer protein could be reached.
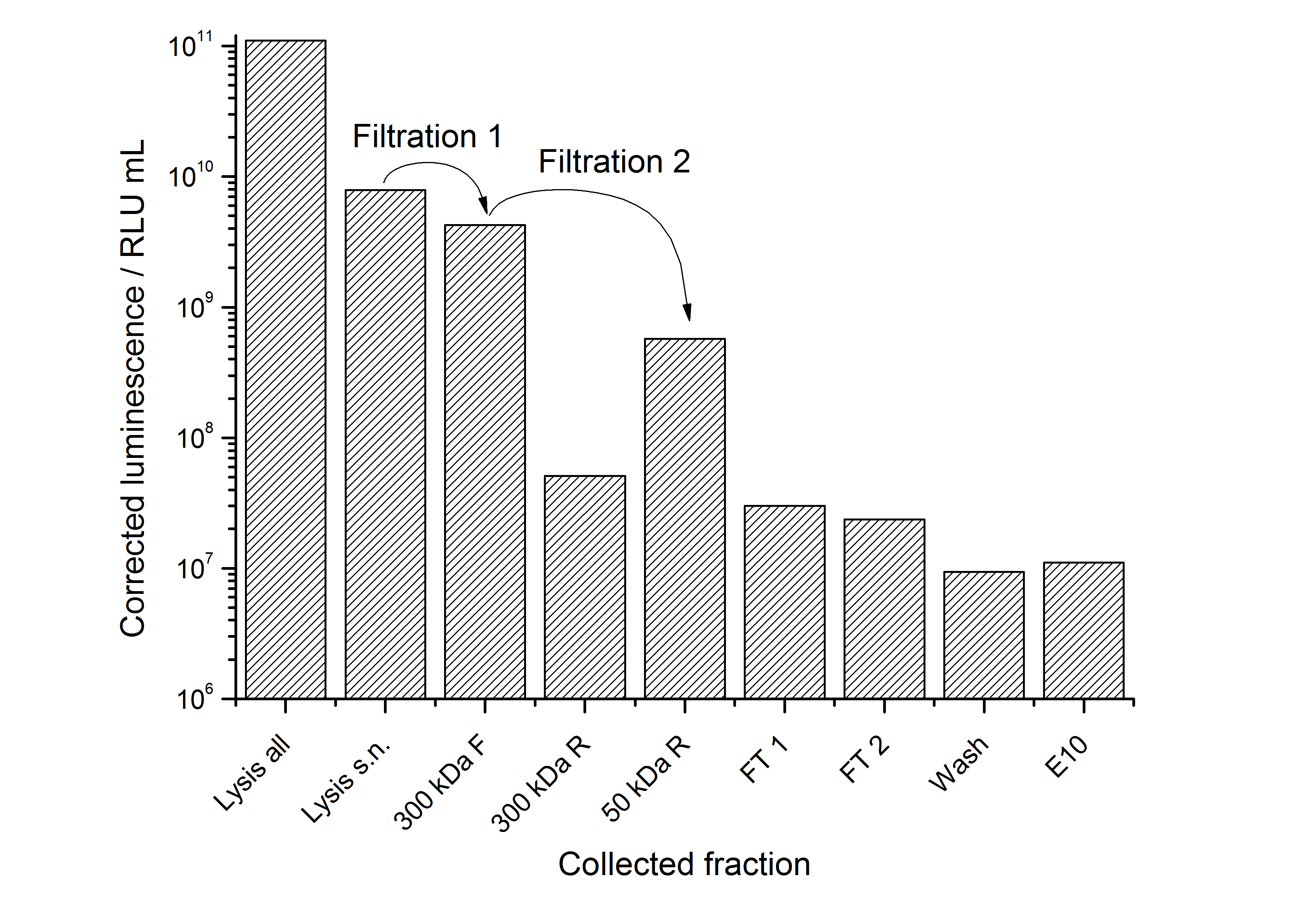
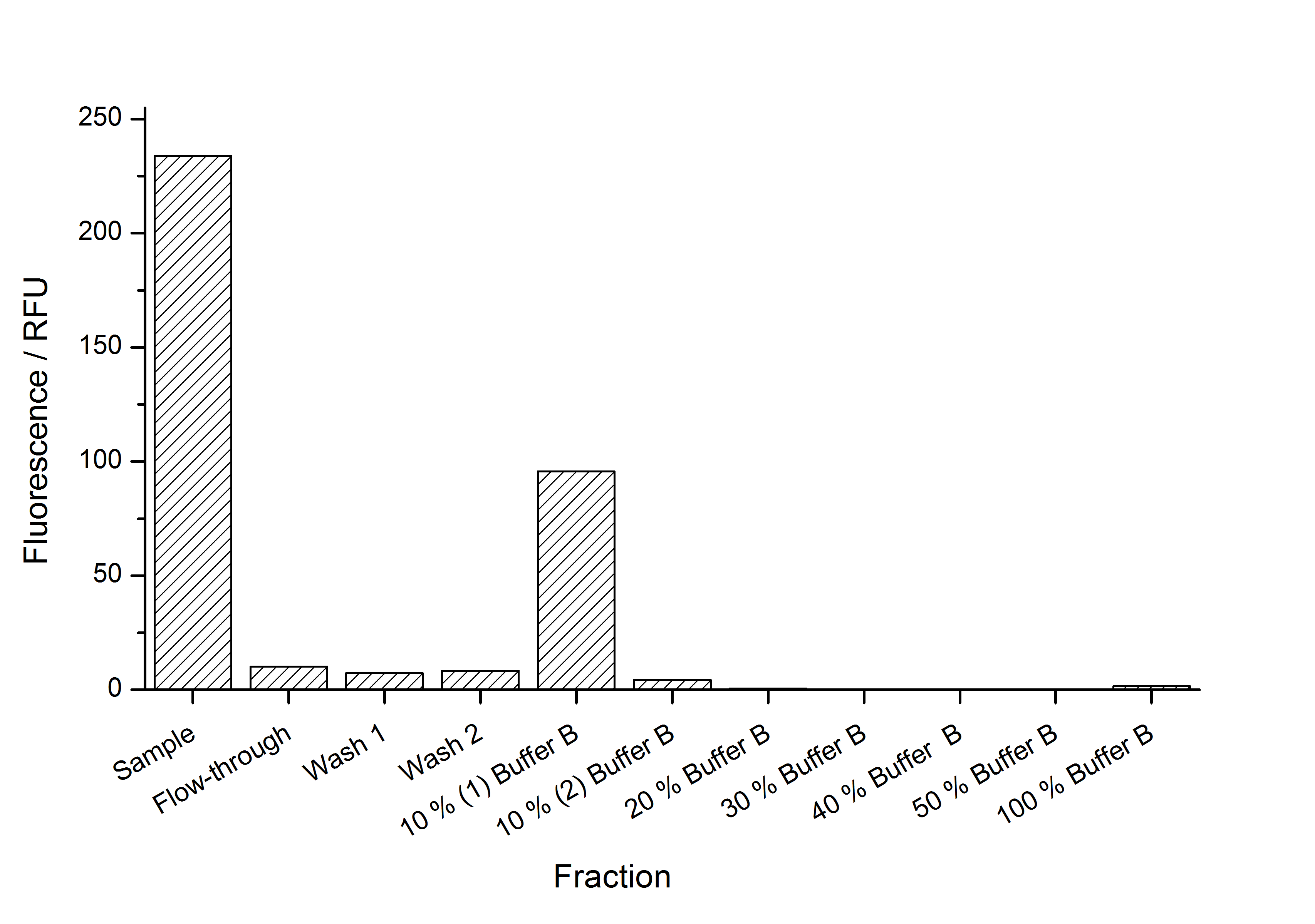
This purification was made using the SgsE fusion protein containing an N-terminal mCitrine for identification and a C-terminal His-6-tag. The SDS-PAGE gel (Figure 11) and the fluorescence (Figure 12) in the collected fractions showed that the majority of fusion protein was eluated with a imidazole concentration of ca. 50 mM. There was also fluorescence measurable in the flowthrough and wash fractions. This indicates that the used 1 mL HisTrap FF crude (GE Healthcare) was overloaded or the protein was bound only weakly to the affinity matrix. The resulting purity in the elution fraction, the saving of time and the easiness makes this procedure to the preferred purification method.

Because the method is so quick and easy now we developed a guide to do it yourself nanobiotechnology in which the method is presented in a vivid way.
Sometimes, the inclusion body purification does not give very clean fractions. Therefore, we developed another purification strategy for <partinfo>K525405</partinfo> monomers after inclusion body purification. This strategy consists of a capture with an IEX followed by a purification with HIC. Because of issues with the PES membranes we used to rebuffer our samples (see below - PES binds most of the S-layers) we lost a lot of our proteins in this step between the two chromatographies. So there was not enough protein left to colour it with Coomassie in an SDS-PAGE and time ran out to perform a silver staining. The fluorescence of the collected fractions of the chromatographies is shown in the figures below.


Enzyme reaction fused to S-layer protein
To test an exemplary enzymatic reaction fused to an S-layer protein SgsE | luciferase was assembled and expressed. The fusion protein shows high luciferase activity inside the cells but unfortunately the purification and stability at room temperature has some issues which have to be solved in future applications.
Expression in E. coli
For characterization experiments the SgsE gene under the control of a T7 / lac promoter (<partinfo>K525303</partinfo>) was fused to firefly luciferase (<partinfo>K525999</partinfo>) using Freiburg BioBrick assembly.
The SgsE | luciferase fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase with 0.2 % L-rhamnose and induction of lac operator with 1 mM IPTG.
The following cultivation was carried out in a [http://www.gmi-inc.com/BioEngineering-KLF-Small-Laboratory-Fermenter.html#product_desc Bioengineering KLF] bioreactor with Bioengineering DCU and software. A sequencer which automatically pumped an inducer solution after 4 h cultivation time to start protein expression was implemented. Other parameters were:
- Medium: HSG medium with 20 mg L-1 chloramphenicol
- Culture volume: 2.5 L
- Starting OD600: 0.4
- DO: 60 % air saturation (controlled with stirrer cascade starting with 200 rpm)
- pH: 7.0 (controlled with 20 % phosphoric acid and 2 M NaOH)
- Antifoam: BASF pluronic PE-8100
- Induction solution: 0.2 % L-rhamnose and 1 mM IPTG
The following figure shows the expression of the SgsE | luciferase S-layer fusion protein (<partinfo>K525311</partinfo>) in E. coli KRX in HSG medium with autoinduction sequencer as described above. Optical density, activity of the fused luciferase, dissolved oxygen and agitator speed are plotted against the cultivation time.

Purification
After the analysis of cultivations with expression of SgsE | luciferase fusion proteins, different cell fractions were analyzed. It could be seen that the proteins form inclusion bodies in E. coli but that there are some soluble proteins left. This has the advantage that the proteins carrying an enzyme as fusion protein do not have to be treated with denaturating agents like urea which destroys the enzyme (data not shown).
To capture the protein from the cell lysate an ion exchange chromatography (IEX) was carried out (binding with pH 7.0, 25 mM NaCl, quaternary amine beads, elution with 100 mM NaCl). A lot of protein was found in the flow-through. When concentrating and rebuffering the proteins with PES (polyethylene sulfone) membranes a lot of protein was lost. The S-layer proteins stuck to the membrane. Some could be removed again from the membrane after cutting out the filter and incubate it in ddH2O over-night. This problem has to be kept in mind when using this S-layer.
The results of the purification approach are shown in Figure 16:
The purification strategy has to be improved. The inclusion bodies cannot be purified because urea damages the luciferase irreversible (data not shown). The loss due to adsorption of the SgsE | luciferase fusion protein to PES membranes could be avoided by using different membranes. The binding conditions of the IEX have to be improved as well. Anyway, the idea behind this purification strategy could be a starting point for a better strategy. Possibilities for improvement are:
- Different membranes for ultra- / diafiltration
- Other binding conditions for the IEX capturing step (higher pH)
- Hydrophobic interaction chromatography as purification step after IEX (works in general, data not shown)
- Size exclusion chromatography (SEC) for polishing
Stability
To test whether the S-layer protein enhances the half-life of the firefly luciferase, immobilization experiments were carried out. After the IEX described above, the elution fraction was immobilized on silicon dioxide beads or just diluted with HBSS buffer which is used for the immobilization / recrystallization of SgsE S-layer proteins. The immobilization is carried out at room temperature for 4 h. It could be seen that the luciferase activity nearly expired during this time (in the positive and the negative control). So the S-layer SgsE could not stabilize the luciferase at room temperature. The results of this experiment is shown in Figure 17:
NAD+ detection
Identification of LigA
The gel bands of SDS-PAGE for His-tag purified LigA were analysed by MALDI-TOF MS and the comparison with the Swiss-Prot database clearly identified the purified protein as LigA (Figure 18, Table 2). As it is reported in literature the majority of LigA usually occurs in its adenylated form after extraction from E. coli. An indication for this is the double band on the right edge of Figure 18. The proteins in both gel bands were identified as LigA from E. coli suggesting that gel band 3 is the adenylated form and gel band 4 is the deadenylated form.
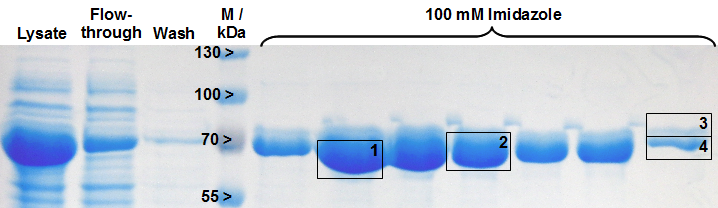
Table 2: Identification of LigA by MALDI-TOF MS. The values correspond to the framed and numbered gel bands in Figure 18. The threshold for significance of the Mascot Score for MS is 63 and the one for MS/MS is 26. The MS-Coverage represents the sequence coverage of the investigated protein with the corresponding entry in the E. coli Swiss-Prot data base. The Sequence-Coverage shows the percentage of similarity to the translated BioBrick <partinfo>BBa_K525710</partinfo> (translation was perfomed with the [http://web.expasy.org/translate/ ExPASy Translate] tool).
| Number | Method | Swiss-Prot number | Protein | Protein Mascot Score | Protein MW | pI-Value | MS- Coverage [%] | Sequence- Coverage [%] |
|---|---|---|---|---|---|---|---|---|
| 1 | MS | B1XA82 | DNA ligase OS=Escherichia coli GN=ligA | 96 | 73560 | 5.3 | 17 | 25.8 |
| 2 | MS | B1XA82 | DNA ligase OS=Escherichia coli GN=ligA | 109 | 73560 | 5.3 | 21 | 28.7 |
| 3 | MS | A7ZPL2 | DNA ligase OS=Escherichia coli GN=ligA | 41 | 73618 | 5.2 | 7 | 10.2 |
| 4 | MS | B1XA82 | DNA ligase OS=Escherichia coli GN=ligA | 134 | 73560 | 5.3 | 29 | 36.6 |
| 1 | MS/MS | Q0TF55 | DNA ligase OS=Escherichia coli GN=ligA | 54 | 73602 | 5.2 | 6 | / |
| 2 | MS/MS | Q0TF55 | DNA ligase OS=Escherichia coli GN=ligA | 63 | 73602 | 5.2 | 6 | / |
| 3 | MS/MS | Q0TF55 | DNA ligase OS=Escherichia coli GN=ligA | 33 | 73602 | 5.2 | 2 | / |
| 4 | MS/MS | Q0TF55 | DNA ligase OS=Escherichia coli GN=ligA | 36 | 73602 | 5.2 | 2 | / |
Limit of detection
To determine the limit of detection of the NAD+ bioassay the NAD+ concentrations were decreased until the fluorescence enhancement was no longer distinguishable from added H2O. This was the case as soon as the NAD+ concentration came below 2 nM. Deductively, the limit of detection of the molecular beacon based and LigA associated NAD+ bioassay was set at 2 nM NAD+. Furthermore, this confirmed the linear dependence between the initial velocity and the NAD+ concentration (Figure 19).

Selectivity test
The specifity of LigA for its substrate NAD+ is important in order to couple the NAD+ detection with investigated processes including NADH-dependent enzyme reactions. The verification of the selectivity is an important matter to exclude any unspecific reactions which might result in a NAD+-independent fluorescence enhancement. Hence, a selectivity test for LigA was performed with the analytes NADH, NADP+, NADPH, 3-ADAP (NAD+ with an exchanged functional group at the nicotinamide ring system), ATP and ADP, and the relative fluorescence enhancement rates in a NAD+ bioassay were compared with the one for NAD+ (Figure 20).
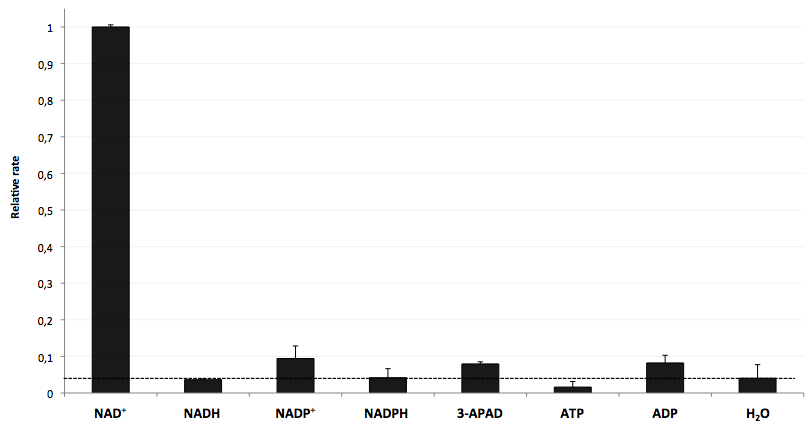
The negative control (H2O) shows that there was a small background signal about 5 % of the signal that was produced by NAD+ when using 100 nM of analytes. This marks the threshold for fluorescence enhancement which is caused by the employed analyte. NADH, NADPH and ATP were similar to the negative control and can thereby seen as analytes that do not enable DNA ligation by LigA. Only the values for the three analytes NADP+, 3-APAD and ADP were above the threshold, but they were constantly below 10 % of the NAD+ signal. This leads to the suggestion that LigA ist highly selective for NAD+ even in presence of structurally similar analytes. This makes the NAD+ bioassay and the associated enzyme LigA suitable for investigating NADH-dependent enzyme reactions or measuring NAD+ in biological analyte mixtures such as cell lysates.
Coupled enzyme reaction
The coupling of the LigA-including NAD+ detection system was performed with a NADH-dependent enzymatic reaction which was in concrete the conversion of pyruvate to L-lactate by lactic acid dehydrogenase (LDH) from E. coli. For this the LDH reaction was done with an excess of NADH and various pyruvate concentrations. Afterwards, the reaction mix was transferred to a NAD+ bioassay and the fluorescence intensity was monitored. In Figure 21 the normalized initial fluorescence enhancement rates for the employed pyruvate concentrations are indicated. The calculated initial velocity was then plotted against the pyruvate concentration (Figure 22).
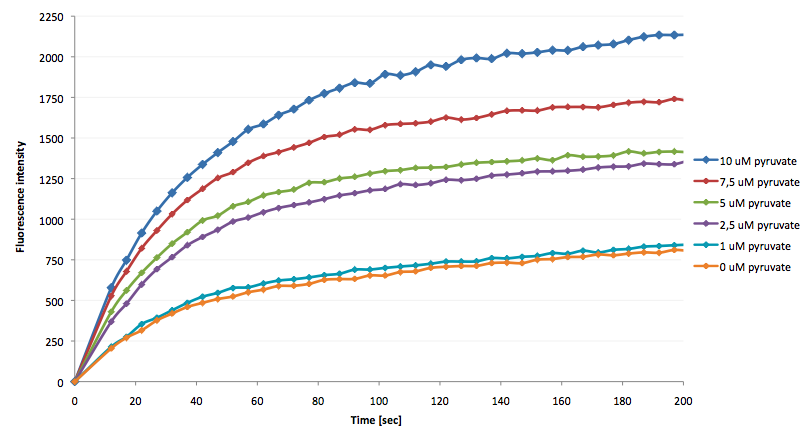
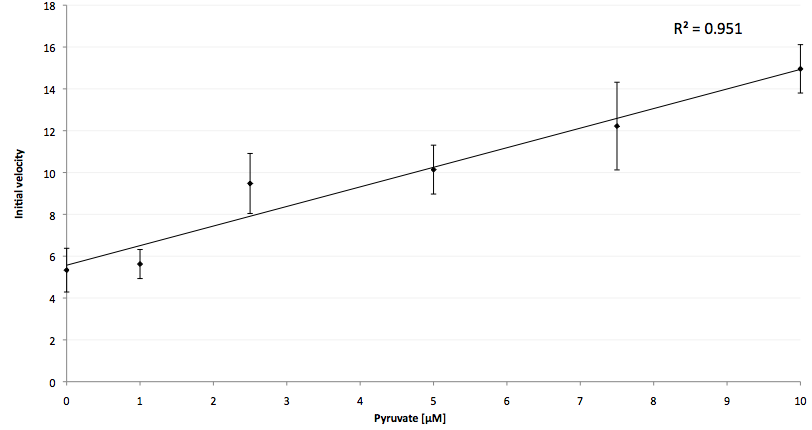
The addition of the LDH reaction mix resulted in a characteristic fluorescence enhancement rate depending on the employed pyruvate concentration. The existing correlation between both parameters seemed to be a linear. That the signal for 0 µM pyruvate was remarkably high could be the result of an pyruvate-independent transfer of electrons from NADH to the active site histidine of LDH under formation of NAD+. The limit of detection for pyruvate seemed to be near 1 µM pyruvate. That this value was not in nano molarity scale is firstly caused by 100-fold dilution of the LDH reaction mix after addition to the NAD+ bioassay and secondly by the fact that LDH does not necessarily converte 100 % of the pyruvate to L-lactate. Nevertheless, for the LigA based NAD+ detection system it has been proven that it can be coupled to NADH-dependent enzymatic reactions. This makes the NAD+ bioassay suitable for a wide range of biological studies dealing with the ubiquitous cofactors NADH/NAD+.
Molecular cloning
Because DNA ligase from E. coli is commercially aquirable for cloning purposes LigA (<partinfo>BBa_K525710</partinfo>) was tested whether it is also suitable for molecular cloning procedures. Hence, cloning of rfp into a pSB1C3 vector was performed with LigA. Ligation was done with NAD+ bioassay buffer containing 10 mM NAD+ at 37 °C or 22 °C and the vectors were transformed into E. coli KRX by electroporation. After growth over night at 37 °C on petri dishes the results were documented (Figure 23).
By analyzing the number of colony forming units and color of the colonies the ligation efficiency of LigA should be assessable. As shown for the sample without any ligase only few clones were able to grow on chloramphenicol supplemented LB medium. A much bigger number of clones was observed when ligation by LigA was performed at 22 °C. Furthermore, there were a lot of red colonies indicating positive clones. Deductively, LigA might be useful for (large-scale) molecular cloning procedures which could be beneficial because of its easy production in E. coli and simple purification.
 "
"