Team:SJTU-BioX-Shanghai/Parts
From 2011.igem.org
(→BBa_K567013 tRNAAsp-TAG(Favorite Part)) |
(→SJTU-BioX-Shanghai 2011 iGEM Team Parts) |
||
| (46 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
====SJTU-BioX-Shanghai 2011 iGEM Team Parts==== | ====SJTU-BioX-Shanghai 2011 iGEM Team Parts==== | ||
| - | {|style="background-color:#fff; border-bottom: | + | {|style="background-color:#fff; border-bottom:3px solid #1192b9;" |
|-style="background-color:#c8e2ea;" | |-style="background-color:#c8e2ea;" | ||
!colspan="2" width="60"| -?- | !colspan="2" width="60"| -?- | ||
| Line 91: | Line 91: | ||
|[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567002 BBa_K567002] | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567002 BBa_K567002] | ||
|Translational_Unit | |Translational_Unit | ||
| - | |[[# | + | |[[#BBa_K567002_sulA_promoter-tRNAArg|''sulA'' promoter-tRNA<sup>Arg</sup>]] |
|Yunfeng Ruan and Chunying Li | |Yunfeng Ruan and Chunying Li | ||
|567 | |567 | ||
| Line 199: | Line 199: | ||
|[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567020 BBa_K567020] | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567020 BBa_K567020] | ||
|Reporter | |Reporter | ||
| - | | | + | |KanR |
|Zhang Kaisi | |Zhang Kaisi | ||
|816 | |816 | ||
| + | |||
| + | |- | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567021 BBa_K567021] | ||
| + | |Reporter | ||
| + | |[[#BBa_K567021_PT7-Luc-2x4AGG|PT7-Luc-2x4AGG]] | ||
| + | |Wang You | ||
| + | |1885 | ||
| + | |||
| + | |-style="background-color:#ddf3ff;" | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567022 BBa_K567022] | ||
| + | |Translational_Unit | ||
| + | |[[#BBa_K567022_PT7-Luc-2AGG-tRNAArg|PT7-Luc-2AGG-tRNA(Arg)]] | ||
| + | |You Wang | ||
| + | |2037 | ||
| + | |||
| + | |-style="background-color:#ddf3ff;" | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567023 BBa_K567023] | ||
| + | |Translational_Unit | ||
| + | |[[#BBa_K567022_PT7-Luc-2AGG-tRNAArg|PT7-Luc-4AGG-tRNA(Arg)]] | ||
| + | |You Wang | ||
| + | |2043 | ||
| + | |||
| + | |- | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567024 BBa_K567024] | ||
| + | |Translational_Unit | ||
| + | |[[#BBa_K567022_PT7-Luc-2AGG-tRNAArg|PT7-Luc-6AGG-tRNA(Arg)]] | ||
| + | |You Wang | ||
| + | |2049 | ||
| + | |||
| + | |-style="background-color:#ddf3ff;" | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567025 BBa_K567025] | ||
| + | |Translational_Unit | ||
| + | |[[#BBa_K567022_PT7-Luc-2AGG-tRNAArg|PT7-Luc-8AGG-tRNA(Arg)]] | ||
| + | |You Wang | ||
| + | |2055 | ||
| + | |||
| + | |- | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567026 BBa_K567026] | ||
| + | |Reporter | ||
| + | |[[#BBa_K567026_PT7-Luc-2x4AGG.2830bp.29|PT7-Luc-2x4AGG(30bp)]] | ||
| + | |You Wang | ||
| + | |1798 | ||
| + | |||
| + | |-style="background-color:#ddf3ff;" | ||
| + | | | ||
| + | |W | ||
| + | |[http://partsregistry.org/wiki/index.php?title=Part:BBa_K567027 BBa_K567027] | ||
| + | |Reporter | ||
| + | |[[#BBa_K567027_PT7-Luc-3x4AGG.2830bp.2B111bp.29|PT7-Luc-3x4AGG(30bp+111bp)]] | ||
| + | |You Wang | ||
| + | |1936 | ||
|} | |} | ||
| - | [[image:11SJTU_Parts_favor.png]]: Favorite Part; '''W''':Works | + | '''Note''': [[image:11SJTU_Parts_favor.png]]: Favorite Part; '''W''':Works |
Click '''Name''' to the Partsregistry Main Page, Click '''Description''' to see construction information. | Click '''Name''' to the Partsregistry Main Page, Click '''Description''' to see construction information. | ||
| - | ===1. | + | ===1.Codon Switch=== |
| - | |||
| - | ====1.1 tRNA | + | ====1.1 Rare-Codon Switch: control with rare tRNA amount:==== |
[[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| Line 223: | Line 285: | ||
[[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| - | =====BBa_K567002 | + | =====BBa_K567002 ''sulA'' promoter-tRNA<sup>Arg</sup>===== |
Description: ''sulA'' promoter derived from ''E.coli'' ''sulA'' operon and tRNA<sup>Arg</sup> from ''E.coli'' ''ArgW'' operon. The tRNA<sup>Arg</sup> is under the control of ''sulA'' promoter induced by UV. tRNA expression is induced by 20-second UV exposure (the distance between the 20W lamp and the culture is 35cm) when the OD600 of the culture reaches 0.3. This part is constructed on the backbone plasmid pACYC184. | Description: ''sulA'' promoter derived from ''E.coli'' ''sulA'' operon and tRNA<sup>Arg</sup> from ''E.coli'' ''ArgW'' operon. The tRNA<sup>Arg</sup> is under the control of ''sulA'' promoter induced by UV. tRNA expression is induced by 20-second UV exposure (the distance between the 20W lamp and the culture is 35cm) when the OD600 of the culture reaches 0.3. This part is constructed on the backbone plasmid pACYC184. | ||
| Line 229: | Line 291: | ||
[[image:11SJTU-UV.jpg|500px|''sulA'' promoter-tRNA(Arg)]] | [[image:11SJTU-UV.jpg|500px|''sulA'' promoter-tRNA(Arg)]] | ||
| - | ====1.2 | + | ====1.2 Rare-Codon Switch: control with aaRS ==== |
[[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| Line 235: | Line 297: | ||
Description: tRNA<sup>Asp</sup> with its anticodon mutated to CCU(base pairing rare codon AGG) and under the control of ''aspV'' promoter. This biobrick is constructed first by cloning the tRNA<sup>Asp</sup> from ''AspV'' in ''E.coli'', then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. | Description: tRNA<sup>Asp</sup> with its anticodon mutated to CCU(base pairing rare codon AGG) and under the control of ''aspV'' promoter. This biobrick is constructed first by cloning the tRNA<sup>Asp</sup> from ''AspV'' in ''E.coli'', then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. | ||
| - | [[image:11SJTU-tRNA-asp-AGG.jpg|tRNA<sup>Asp</sup>-AGG]] | + | [[image:11SJTU-tRNA-asp-AGG.jpg|500px|tRNA<sup>Asp</sup>-AGG]] |
[[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| Line 243: | Line 305: | ||
Description: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and ''lac'' operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from ''E.coli''. This modified AspRS can charge Asp to tRNA<sup>Asp</sup>-TAG (BBa_K567013) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene ''AspS'' into the multiple cloning site on the plasmid pET28a. | Description: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and ''lac'' operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from ''E.coli''. This modified AspRS can charge Asp to tRNA<sup>Asp</sup>-TAG (BBa_K567013) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene ''AspS'' into the multiple cloning site on the plasmid pET28a. | ||
| - | ====1.3 Stop-Codon | + | ====1.3 Stop-Codon Switch: ==== |
[[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| Line 249: | Line 311: | ||
Description: tRNA<sup>Asp</sup> with its anticodon mutated to CUA (base pairing stop codon UAG) and under the control of ''aspV'' promoter. This biobrick is constructed first by cloning the tRNA<sup>Asp</sup> from ''AspV'' in ''E.coli'', then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. | Description: tRNA<sup>Asp</sup> with its anticodon mutated to CUA (base pairing stop codon UAG) and under the control of ''aspV'' promoter. This biobrick is constructed first by cloning the tRNA<sup>Asp</sup> from ''AspV'' in ''E.coli'', then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. | ||
| - | [[image:11SJTU-tRNA-asp-TAG.jpg|tRNA<sup>Asp</sup>-TAG]] | + | [[image:11SJTU-tRNA-asp-TAG.jpg|500px|tRNA<sup>Asp</sup>-TAG]] |
| + | |||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567011 PT7-TDRS===== | =====BBa_K567011 PT7-TDRS===== | ||
| Line 255: | Line 319: | ||
Description: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and ''lac'' operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from ''E.coli''. This modified AspRS can charge Asp to tRNA<sup>Asp</sup>-TAG (BBa_K567013) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene ''AspS'' into the multiple cloning site on the plasmid pET28a. | Description: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and ''lac'' operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from ''E.coli''. This modified AspRS can charge Asp to tRNA<sup>Asp</sup>-TAG (BBa_K567013) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene ''AspS'' into the multiple cloning site on the plasmid pET28a. | ||
| - | ====1.4 Initial-Codon | + | ====1.4 Initial-Codon Switch:==== |
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567014 PT7-''metG''M===== | =====BBa_K567014 PT7-''metG''M===== | ||
Description: T7 promoter-''metG''(mutated). This biobrick is constructed by putting the mutated metG (Met-RS) under the control of T7 promoter and ''lac'' operator. We have cloned ''metG'' from ''E.coli'' and have used error-prone PCR to amplify the ''metG''. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and ''metY''-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the Gene ''metG'' fragment with random mutation into the multiple cloning site on the plasmid pET28a. | Description: T7 promoter-''metG''(mutated). This biobrick is constructed by putting the mutated metG (Met-RS) under the control of T7 promoter and ''lac'' operator. We have cloned ''metG'' from ''E.coli'' and have used error-prone PCR to amplify the ''metG''. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and ''metY''-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the Gene ''metG'' fragment with random mutation into the multiple cloning site on the plasmid pET28a. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567015 PT7-''metG''N===== | =====BBa_K567015 PT7-''metG''N===== | ||
Description: T7 promoter-''metG''(truncated). This biobrick is constructed by putting the truncated metG (Met-RS) under the control of T7 promoter and ''lac'' operator. We have cloned ''metG'' from ''E.coli'' and the tRNA recognition domain of metG is truncated. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and ''metY''-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the truncated Gene ''metG'' fragment into the multiple cloning site on the plasmid pET28a. | Description: T7 promoter-''metG''(truncated). This biobrick is constructed by putting the truncated metG (Met-RS) under the control of T7 promoter and ''lac'' operator. We have cloned ''metG'' from ''E.coli'' and the tRNA recognition domain of metG is truncated. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and ''metY''-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the truncated Gene ''metG'' fragment into the multiple cloning site on the plasmid pET28a. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567016 ''metY''-CGA===== | =====BBa_K567016 ''metY''-CGA===== | ||
Description: This biobrick is constructed by mutating the anticodon of tRNA<sup>met</sup> to TCG (base pairing codon CGA). This tRNA can transfer fMet to CGA when it is used as the start codon. Kana gene with start codon substituted for CGA is used to testify the function of ''metY''-CGA. When this biobrick and ''metG''M(BBa_K567014) or ''metG''N(BBa_K567015) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on the backbone plasmid pACYC184. | Description: This biobrick is constructed by mutating the anticodon of tRNA<sup>met</sup> to TCG (base pairing codon CGA). This tRNA can transfer fMet to CGA when it is used as the start codon. Kana gene with start codon substituted for CGA is used to testify the function of ''metY''-CGA. When this biobrick and ''metG''M(BBa_K567014) or ''metG''N(BBa_K567015) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on the backbone plasmid pACYC184. | ||
| Line 271: | Line 338: | ||
---- | ---- | ||
| - | + | ===2.Reporter=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
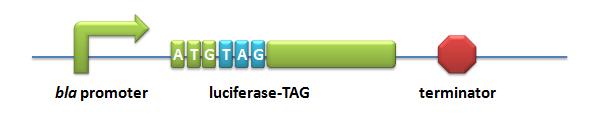
=====BBa_K567003 P''bla''-Luc-TAG===== | =====BBa_K567003 P''bla''-Luc-TAG===== | ||
| Line 283: | Line 347: | ||
[[image:11SJTU-LUC-TAG.jpg|Pbla-Luc-TAG]] | [[image:11SJTU-LUC-TAG.jpg|Pbla-Luc-TAG]] | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567004 P''bla''-Luc-2AGG===== | =====BBa_K567004 P''bla''-Luc-2AGG===== | ||
| Line 297: | Line 362: | ||
Point mutation is used to obtain this part from wild type. '''BBa_K567005 P''bla''-Luc-4AGG, BBa_K567006 P''bla''-Luc-6AGG and BBa_K567007 P''bla''-Luc-8AGG''' are constructed in the same way. | Point mutation is used to obtain this part from wild type. '''BBa_K567005 P''bla''-Luc-4AGG, BBa_K567006 P''bla''-Luc-6AGG and BBa_K567007 P''bla''-Luc-8AGG''' are constructed in the same way. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567005 P''bla''-Luc-4AGG===== | =====BBa_K567005 P''bla''-Luc-4AGG===== | ||
Description: β-lactamase promoter-Luciferase with four AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567005 P''bla''-Luc-4AGG is constructed in the same way as '''BBa_K567004 P''bla''-Luc-2AGG'''. | Description: β-lactamase promoter-Luciferase with four AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567005 P''bla''-Luc-4AGG is constructed in the same way as '''BBa_K567004 P''bla''-Luc-2AGG'''. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567006 P''bla''-Luc-6AGG===== | =====BBa_K567006 P''bla''-Luc-6AGG===== | ||
Description: β-lactamase promoter-Luciferase with six AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567006 P''bla''-Luc-6AGG is constructed in the same way as '''BBa_K567004 P''bla''-Luc-2AGG'''. | Description: β-lactamase promoter-Luciferase with six AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567006 P''bla''-Luc-6AGG is constructed in the same way as '''BBa_K567004 P''bla''-Luc-2AGG'''. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567007 P''bla''-Luc-8AGG===== | =====BBa_K567007 P''bla''-Luc-8AGG===== | ||
| Line 311: | Line 379: | ||
[[image:11SJTU-bla-AGG-luc.jpg|600px|Pbla-Luc]] | [[image:11SJTU-bla-AGG-luc.jpg|600px|Pbla-Luc]] | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
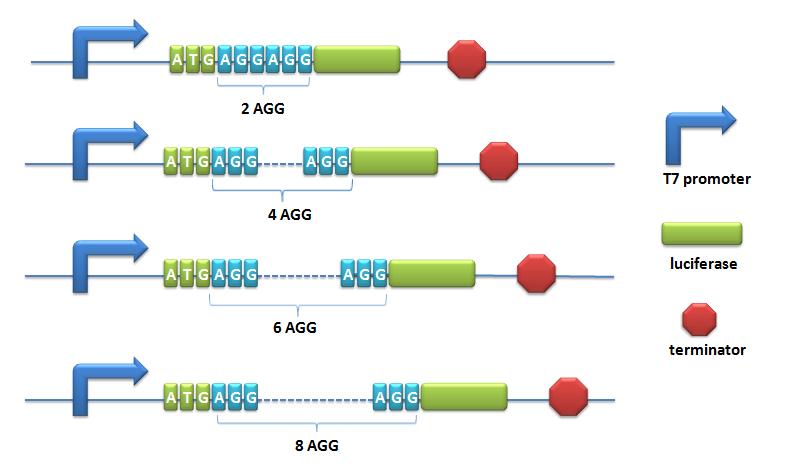
=====BBa_K567008 PT7-Luc-2AGG===== | =====BBa_K567008 PT7-Luc-2AGG===== | ||
Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 2 AGG codons and 2 GCG codons are inserted after the ATG start codon of wild type luciferase (BBa_I712019). Modified luciferase keeps the activity of converting luciferin into oxyluciferin, during which bioluminescence will emit. This part is one of the reporter genes to testify the influence of different number of rare codons in regulating protein biosynthesis. This part is used as a measurement to testify the function of ''LacI'' -Ptrc-tRNA<sup>Arg</sup> (BBa_K567001) or ''sulA'' promoter-tRNA<sup>Arg</sup> (BBa_K567002). | Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 2 AGG codons and 2 GCG codons are inserted after the ATG start codon of wild type luciferase (BBa_I712019). Modified luciferase keeps the activity of converting luciferin into oxyluciferin, during which bioluminescence will emit. This part is one of the reporter genes to testify the influence of different number of rare codons in regulating protein biosynthesis. This part is used as a measurement to testify the function of ''LacI'' -Ptrc-tRNA<sup>Arg</sup> (BBa_K567001) or ''sulA'' promoter-tRNA<sup>Arg</sup> (BBa_K567002). | ||
| Line 324: | Line 393: | ||
'''BBa_K567009 PT7-Luc-4AGG, BBa_K567010 PT7-Luc-8AGG and BBa_K567019 PT7-Luc-6AGG''' are constructed in the same way. | '''BBa_K567009 PT7-Luc-4AGG, BBa_K567010 PT7-Luc-8AGG and BBa_K567019 PT7-Luc-6AGG''' are constructed in the same way. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567009 PT7-Luc-4AGG===== | =====BBa_K567009 PT7-Luc-4AGG===== | ||
Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567019 PT7-Luc-6AGG===== | =====BBa_K567019 PT7-Luc-6AGG===== | ||
Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
=====BBa_K567010 PT7-Luc-8AGG===== | =====BBa_K567010 PT7-Luc-8AGG===== | ||
Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | Description: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as '''BBa_K567008 PT7-Luc-2AGG'''. | ||
| Line 335: | Line 407: | ||
[[image:11SJTU-T7-AGG-luc.jpg|600px|PT7-Luc]] | [[image:11SJTU-T7-AGG-luc.jpg|600px|PT7-Luc]] | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
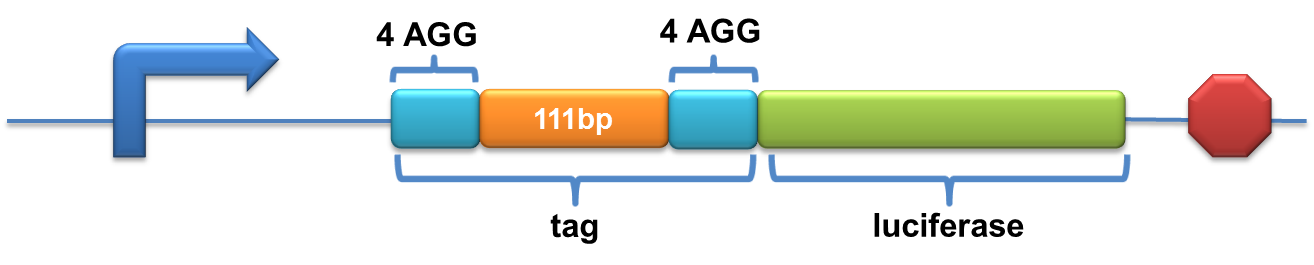
| + | =====BBa_K567021 PT7-Luc-2x4AGG(111bp)===== | ||
| + | Description: This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in TWO DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an inetrval of 111bp. | ||
| + | |||
| + | [[image:11SJTU-2X4AGG-111bp.png|600px]] | ||
| + | |||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
| - | |||
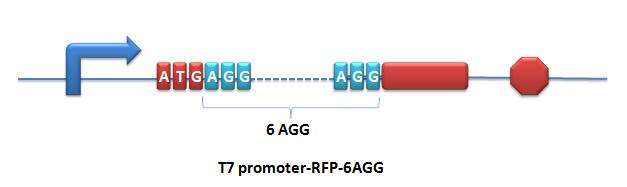
=====BBa_K567017 PT7-RFP-6AGG===== | =====BBa_K567017 PT7-RFP-6AGG===== | ||
Description: RFP with 6 AGG-codon insertions after ATG under the control of T7 promoter and lac operator. The protein can be expressed successfully when co-transformed with PT7-TDRS (BBa_K567011) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). Modified RFP exhibits lower red fluorescence brightness. This part is used to testify the function of PT7-TDRS (BBa_K567011). This part is constructed by inserting the RFP-6AGG fragment into the multiple cloning site on the plasmid pET28a. | Description: RFP with 6 AGG-codon insertions after ATG under the control of T7 promoter and lac operator. The protein can be expressed successfully when co-transformed with PT7-TDRS (BBa_K567011) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). Modified RFP exhibits lower red fluorescence brightness. This part is used to testify the function of PT7-TDRS (BBa_K567011). This part is constructed by inserting the RFP-6AGG fragment into the multiple cloning site on the plasmid pET28a. | ||
| Line 343: | Line 421: | ||
[[image:11SJTU-RFP-6AGG.jpg|PT7-RFP-6AGG]] | [[image:11SJTU-RFP-6AGG.jpg|PT7-RFP-6AGG]] | ||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
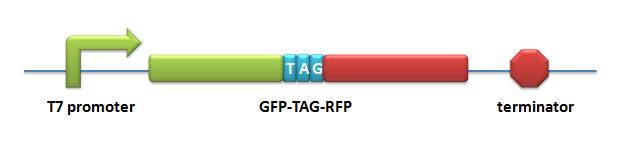
=====BBa_K567018 PT7-GFP-TAG-RFP===== | =====BBa_K567018 PT7-GFP-TAG-RFP===== | ||
Description: GFP and RFP linked with a flexible chain and a stop codon TAG is inserted in the flexible chain. This biobrick is under the control of T7 promoter and lac operator. This part is used to testify the function of PT7-TDRS (BBa_K567011) and tRNA<sup>Asp</sup>-TAG (BBa_K567013). This part is constructed by inserting the GFP-TAG-RFP fragment into the multiple cloning site on the plasmid pET28a. | Description: GFP and RFP linked with a flexible chain and a stop codon TAG is inserted in the flexible chain. This biobrick is under the control of T7 promoter and lac operator. This part is used to testify the function of PT7-TDRS (BBa_K567011) and tRNA<sup>Asp</sup>-TAG (BBa_K567013). This part is constructed by inserting the GFP-TAG-RFP fragment into the multiple cloning site on the plasmid pET28a. | ||
| Line 348: | Line 427: | ||
[[image:11SJTU-GFP-TAG-RFP.jpg|PT7-GFP-TAG-RFP]] | [[image:11SJTU-GFP-TAG-RFP.jpg|PT7-GFP-TAG-RFP]] | ||
| + | |||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
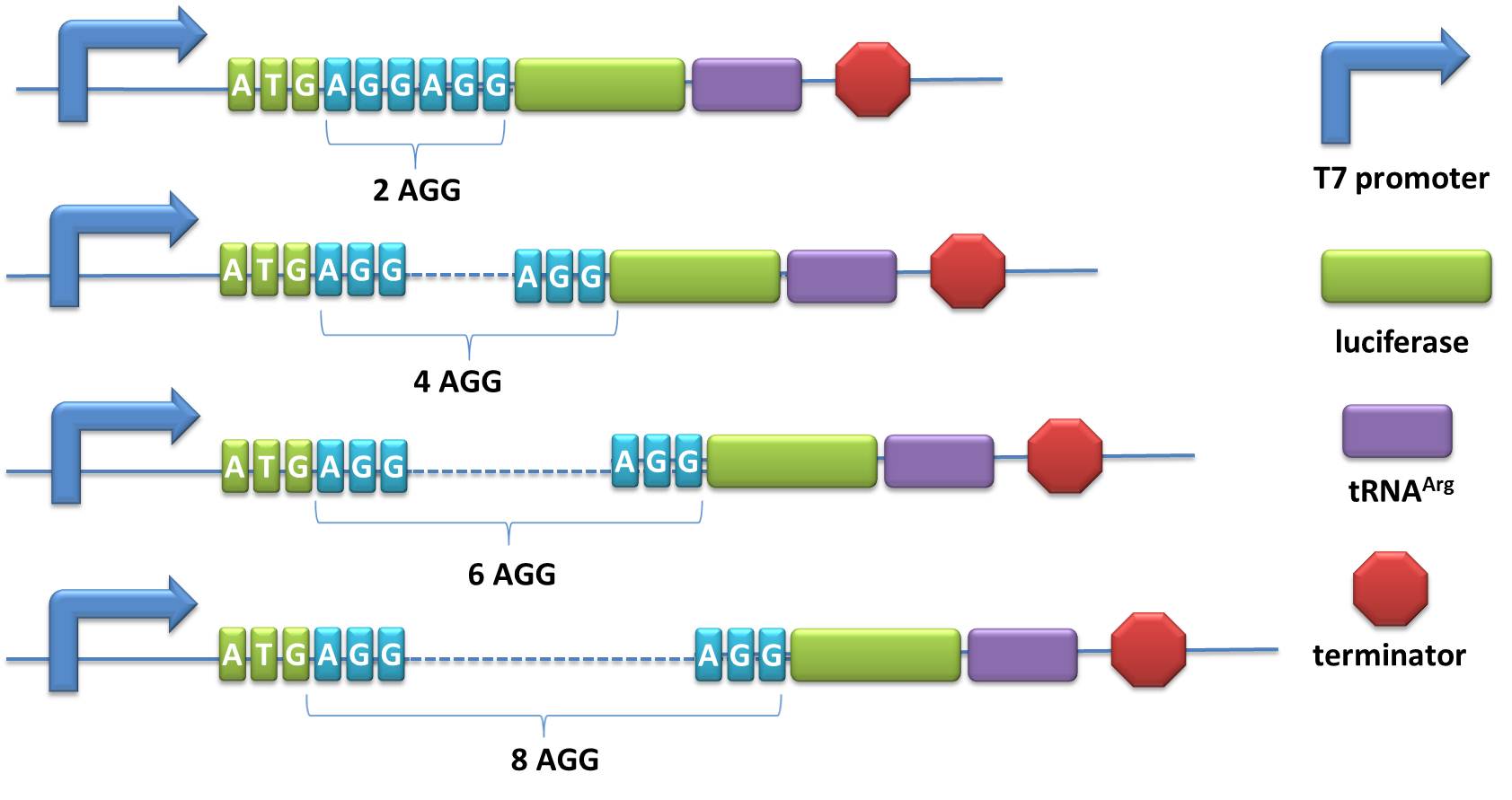
| + | =====BBa_K567022 PT7-Luc-2AGG-tRNA<sup>Arg</sup>===== | ||
| + | Description:This part is constructed based on PT7-Luc-2AGG ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567008 BBa_K567008]). The tDNA<sup>Arg</sup> and luciferase gene into T7 operon so that both genes can be transcribed. tRNA<sup>Arg</sup> can be correctly processed and become matured. Luciferase mRNA will be further translated into protein. The induced tRNA<sup>Arg</sup> can facilitate the translation of luciferase-2AGG, achieving positive feedback. | ||
| + | The transcription of this part is under the control of T7 promoter and ''lac'' operator. RNA expression is induced by 0.5mM IPTG when the OD600 of the culture reaches 0.3. | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567023 BBa_K567023], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567024 BBa_K567024] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567025 BBa_K567025] are constructed in the same way only with different number of AGGs. | ||
| + | |||
| + | [[image:11SJTU-rare codon-tRNA-part.jpg|PT7-Luc-2AGG-tRNA<sup>Arg</sup>|600px]] | ||
| + | |||
| + | |||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
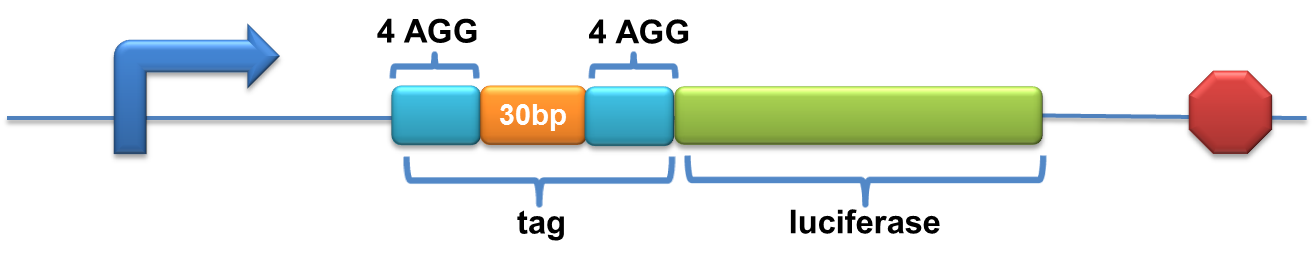
| + | =====BBa_K567026 PT7-Luc-2x4AGG(30bp)===== | ||
| + | Description:This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in TWO DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an interval of 30bp. | ||
| + | |||
| + | [[image:11SJTU-2X4AGG-30bp.png|600px]] | ||
| + | |||
| + | |||
| + | [[image:11SJTU_arrow_top.jpg|right|50px|link=#SJTU-BioX-Shanghai_2011_iGEM_Team_Parts]] | ||
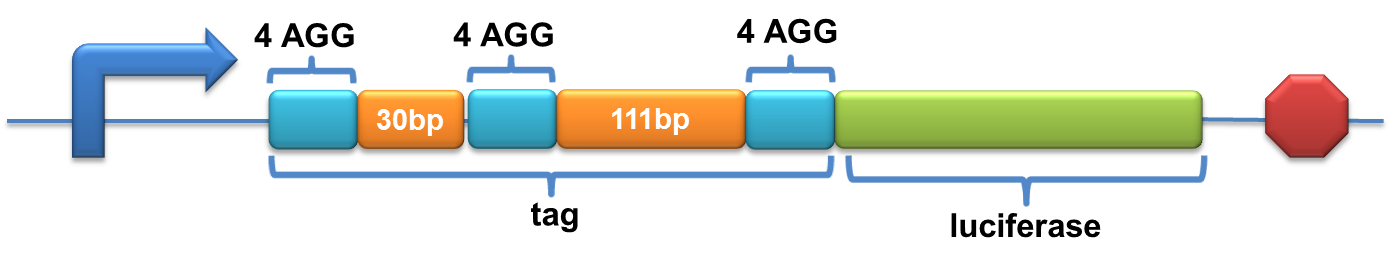
| + | =====BBa_K567027 PT7-Luc-3x4AGG(30bp+111bp)===== | ||
| + | Description:This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in THREE DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an interval of 30bp and 111bp respectively. | ||
| + | |||
| + | [[image:11SJTU-3X4AGG.png|600px]] | ||
[[image:11SJTU_arrow_next.jpg|right|Next Page|link=Team:SJTU-BioX-Shanghai/Parts/Data_Page]] | [[image:11SJTU_arrow_next.jpg|right|Next Page|link=Team:SJTU-BioX-Shanghai/Parts/Data_Page]] | ||
Latest revision as of 03:48, 29 October 2011
|
|
BioBricksSJTU-BioX-Shanghai 2011 iGEM Team Parts
Note: Click Name to the Partsregistry Main Page, Click Description to see construction information. 1.Codon Switch1.1 Rare-Codon Switch: control with rare tRNA amount:BBa_K567001 lacI-Ptrc-tRNAArg(Favorite Part)Description: lacI-Ptrc derived from plasmid pTrc99B and tRNAArg from E.coli ArgW operon. The tRNAArg is under the control of promoter trc. tRNAArg expression is induced by 0.5mM IPTG when the OD600 of the culture reaches 0.3. This part is constructed on the backbone plasmid pACYC184. BBa_K567002 sulA promoter-tRNAArgDescription: sulA promoter derived from E.coli sulA operon and tRNAArg from E.coli ArgW operon. The tRNAArg is under the control of sulA promoter induced by UV. tRNA expression is induced by 20-second UV exposure (the distance between the 20W lamp and the culture is 35cm) when the OD600 of the culture reaches 0.3. This part is constructed on the backbone plasmid pACYC184. 1.2 Rare-Codon Switch: control with aaRSBBa_K567012 tRNAAsp-AGG(Favorite Part)Description: tRNAAsp with its anticodon mutated to CCU(base pairing rare codon AGG) and under the control of aspV promoter. This biobrick is constructed first by cloning the tRNAAsp from AspV in E.coli, then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. BBa_K567011 PT7-TDRS (Favorite Part)Description: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and lac operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from E.coli. This modified AspRS can charge Asp to tRNAAsp-TAG (BBa_K567013) and tRNAAsp-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene AspS into the multiple cloning site on the plasmid pET28a. 1.3 Stop-Codon Switch:BBa_K567013 tRNAAsp-TAG(Favorite Part)Description: tRNAAsp with its anticodon mutated to CUA (base pairing stop codon UAG) and under the control of aspV promoter. This biobrick is constructed first by cloning the tRNAAsp from AspV in E.coli, then the anticodon region is site-directed mutated. This part is constructed on the backbone plasmid pACYC184. BBa_K567011 PT7-TDRSDescription: aspartyl aminoacyl tRNA synthetase without anticodon recognition domain under the control of T7 promoter and lac operator. This biobrick is constructed by deleting the anticodon recognition domain of AspRS from E.coli. This modified AspRS can charge Asp to tRNAAsp-TAG (BBa_K567013) and tRNAAsp-AGG (BBa_K567012). This part is constructed by inserting the truncated fragment of Gene AspS into the multiple cloning site on the plasmid pET28a. 1.4 Initial-Codon Switch:BBa_K567014 PT7-metGMDescription: T7 promoter-metG(mutated). This biobrick is constructed by putting the mutated metG (Met-RS) under the control of T7 promoter and lac operator. We have cloned metG from E.coli and have used error-prone PCR to amplify the metG. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and metY-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the Gene metG fragment with random mutation into the multiple cloning site on the plasmid pET28a. BBa_K567015 PT7-metGNDescription: T7 promoter-metG(truncated). This biobrick is constructed by putting the truncated metG (Met-RS) under the control of T7 promoter and lac operator. We have cloned metG from E.coli and the tRNA recognition domain of metG is truncated. Kana gene with start codon substituted for CGA is used to testify the function of mutated metG. When this biobrick and metY-CGA (BBa_K567016) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on by inserting the truncated Gene metG fragment into the multiple cloning site on the plasmid pET28a. BBa_K567016 metY-CGADescription: This biobrick is constructed by mutating the anticodon of tRNAmet to TCG (base pairing codon CGA). This tRNA can transfer fMet to CGA when it is used as the start codon. Kana gene with start codon substituted for CGA is used to testify the function of metY-CGA. When this biobrick and metGM(BBa_K567014) or metGN(BBa_K567015) are co-transformed into the cell, the cells can survive on the LBKana plate. This part is constructed on the backbone plasmid pACYC184.
2.ReporterBBa_K567003 Pbla-Luc-TAGDescription: Luciferase under the control of β-lactamase promoter with TAG insertion. A TAG codon is inserted after the initial codon of the gene. This part is used to testify the function of corporation of PT7-TDRS (BBa_K567011) and tRNAAsp-TAG (BBa_K567013). β-lactamase promoter is derived from pUC18 β-lactamase operon. Wild type luciferase from BBa_I712019. This part is constructed on the plasmid pET28a as backbone. BBa_K567004 Pbla-Luc-2AGGDescription: β-lactamase promoter-luciferase with two AGG-codon insertions. This biobrick is constructed by putting modified enzyme luciferase under constituitive promoter β-lactamase promoter. 2 AGG codons and 2 GCG codons are inserted after the ATG start codon of wild type luciferase (BBa_I712019). Modified luciferase keeps the activity of converting luciferin into oxyluciferin, during which bioluminescence will emit. This part is one of the reporter genes to testify the influence of different number of rare codons in regulating protein biosynthesis. This part is used as a measurement to testify the function of LacI -Ptrc-tRNAArg(BBa_K567001) or sulA promoter-tRNAArg (BBa_K567002). Cell is cultured in 50ug/ml kanamycin and 10ug/ml tetracycline LB liquid medium. When the OD600 of the culture reaches 0.3 IPTG is added to make the final concentration 0.5nM to induce the synthesis of tRNA. Ultrasonication is used to release the luciferase from the cell. Sonics ON 3 seconds, OFF 3 seconds, total ultrasonication time 3 minutes. Amount of bioluminescence produced can be detected using luminometer. β-lactamase promoter is derived from pUC18 β-lactamase operon. Wild type luciferase from BBa_I712019. This part is constructed on the plasmid pET28a as backbone. Point mutation is used to obtain this part from wild type. BBa_K567005 Pbla-Luc-4AGG, BBa_K567006 Pbla-Luc-6AGG and BBa_K567007 Pbla-Luc-8AGG are constructed in the same way. BBa_K567005 Pbla-Luc-4AGGDescription: β-lactamase promoter-Luciferase with four AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567005 Pbla-Luc-4AGG is constructed in the same way as BBa_K567004 Pbla-Luc-2AGG. BBa_K567006 Pbla-Luc-6AGGDescription: β-lactamase promoter-Luciferase with six AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567006 Pbla-Luc-6AGG is constructed in the same way as BBa_K567004 Pbla-Luc-2AGG. BBa_K567007 Pbla-Luc-8AGGDescription: β-lactamase promoter-Luciferase with eight AGG-codon insertions. Point mutation is used to obtain this part from wild type. BBa_K567007 Pbla-Luc-8AGG is constructed in the same way as BBa_K567004 Pbla-Luc-2AGG. BBa_K567008 PT7-Luc-2AGGDescription: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 2 AGG codons and 2 GCG codons are inserted after the ATG start codon of wild type luciferase (BBa_I712019). Modified luciferase keeps the activity of converting luciferin into oxyluciferin, during which bioluminescence will emit. This part is one of the reporter genes to testify the influence of different number of rare codons in regulating protein biosynthesis. This part is used as a measurement to testify the function of LacI -Ptrc-tRNAArg (BBa_K567001) or sulA promoter-tRNAArg (BBa_K567002). Cell is cultured in 50ug/ml kanamycin 10ug/ml tetracycline LB liquid medium. When the OD600 of the culture reaches 0.3 IPTG is added to make the final concentration 0.5nM to induce the synthesis of tRNA. Ultrasonication is used to release the luciferase from the cell. Sonics ON 3 seconds, OFF 3 seconds, total ultrasonication time 3minutes. Amount of bioluminescence produced can be detected using luminometer. T7 promoter is derived from pET28a. Wild type Luciferase derived from BBa_I712019. BBa_K567009 PT7-Luc-4AGG, BBa_K567010 PT7-Luc-8AGG and BBa_K567019 PT7-Luc-6AGG are constructed in the same way. BBa_K567009 PT7-Luc-4AGGDescription: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as BBa_K567008 PT7-Luc-2AGG. BBa_K567019 PT7-Luc-6AGGDescription: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as BBa_K567008 PT7-Luc-2AGG. BBa_K567010 PT7-Luc-8AGGDescription: this biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator.This biobrick is constructed in the same way as BBa_K567008 PT7-Luc-2AGG. BBa_K567021 PT7-Luc-2x4AGG(111bp)Description: This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in TWO DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an inetrval of 111bp. BBa_K567017 PT7-RFP-6AGGDescription: RFP with 6 AGG-codon insertions after ATG under the control of T7 promoter and lac operator. The protein can be expressed successfully when co-transformed with PT7-TDRS (BBa_K567011) and tRNAAsp-AGG (BBa_K567012). Modified RFP exhibits lower red fluorescence brightness. This part is used to testify the function of PT7-TDRS (BBa_K567011). This part is constructed by inserting the RFP-6AGG fragment into the multiple cloning site on the plasmid pET28a. BBa_K567018 PT7-GFP-TAG-RFPDescription: GFP and RFP linked with a flexible chain and a stop codon TAG is inserted in the flexible chain. This biobrick is under the control of T7 promoter and lac operator. This part is used to testify the function of PT7-TDRS (BBa_K567011) and tRNAAsp-TAG (BBa_K567013). This part is constructed by inserting the GFP-TAG-RFP fragment into the multiple cloning site on the plasmid pET28a.
BBa_K567022 PT7-Luc-2AGG-tRNAArgDescription:This part is constructed based on PT7-Luc-2AGG ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567008 BBa_K567008]). The tDNAArg and luciferase gene into T7 operon so that both genes can be transcribed. tRNAArg can be correctly processed and become matured. Luciferase mRNA will be further translated into protein. The induced tRNAArg can facilitate the translation of luciferase-2AGG, achieving positive feedback. The transcription of this part is under the control of T7 promoter and lac operator. RNA expression is induced by 0.5mM IPTG when the OD600 of the culture reaches 0.3. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567023 BBa_K567023], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567024 BBa_K567024] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K567025 BBa_K567025] are constructed in the same way only with different number of AGGs.
BBa_K567026 PT7-Luc-2x4AGG(30bp)Description:This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in TWO DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an interval of 30bp.
BBa_K567027 PT7-Luc-3x4AGG(30bp+111bp)Description:This biobrick is constructed by putting modified enzyme luciferase under promoter T7 and controlled by lac operator. 4 AGG codons and 2 GCG codons are inserted in THREE DIFFERENT places after the ATG start codon of wild type luciferase (BBa_I712019) with an interval of 30bp and 111bp respectively. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"