Team:Bielefeld-Germany/Labjournal
From 2011.igem.org
(→Week 18: 29th august - 4th september) |
|||
| (160 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Bielefeld_2011_Header}} | {{Bielefeld_2011_Header}} | ||
| + | <html><img src="https://static.igem.org/mediawiki/2011/9/90/Bielefeld-header-labjournal.png"/><p></p></html> | ||
| - | On this page we summarize the (successful) results and achievements of our teamwork. | + | '''Labjournal:''' On this page we summarize the (successful) results and achievements of our teamwork. |
| - | ==Week 1: 2nd - 8th | + | ==Week 1: 2nd - 8th May== |
| - | + | Bisphenol A: | |
| - | + | ||
* cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic) | * cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic) | ||
| - | * cloning of | + | * cloning of fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter |
* testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | * testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection | ||
* expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] | * expression of the successfully assembled BPA degrading BioBricks in ''E. coli'' [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10] | ||
| - | + | S-layer: | |
| - | + | * successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html ''Corynebacterium glutamicum''] and ''Corynebacterium crenatum'' | |
| - | * successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html '' | + | * successful cloning of the S-layer gene of ''Brevibacterium flavum'' without TAT-sequence |
| - | * successful cloning of the S-layer gene of '' | + | |
| - | + | ||
Organizational: | Organizational: | ||
* meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld | * meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld | ||
| - | ==Week 2: 9th - 15th | + | ==Week 2: 9th - 15th May== |
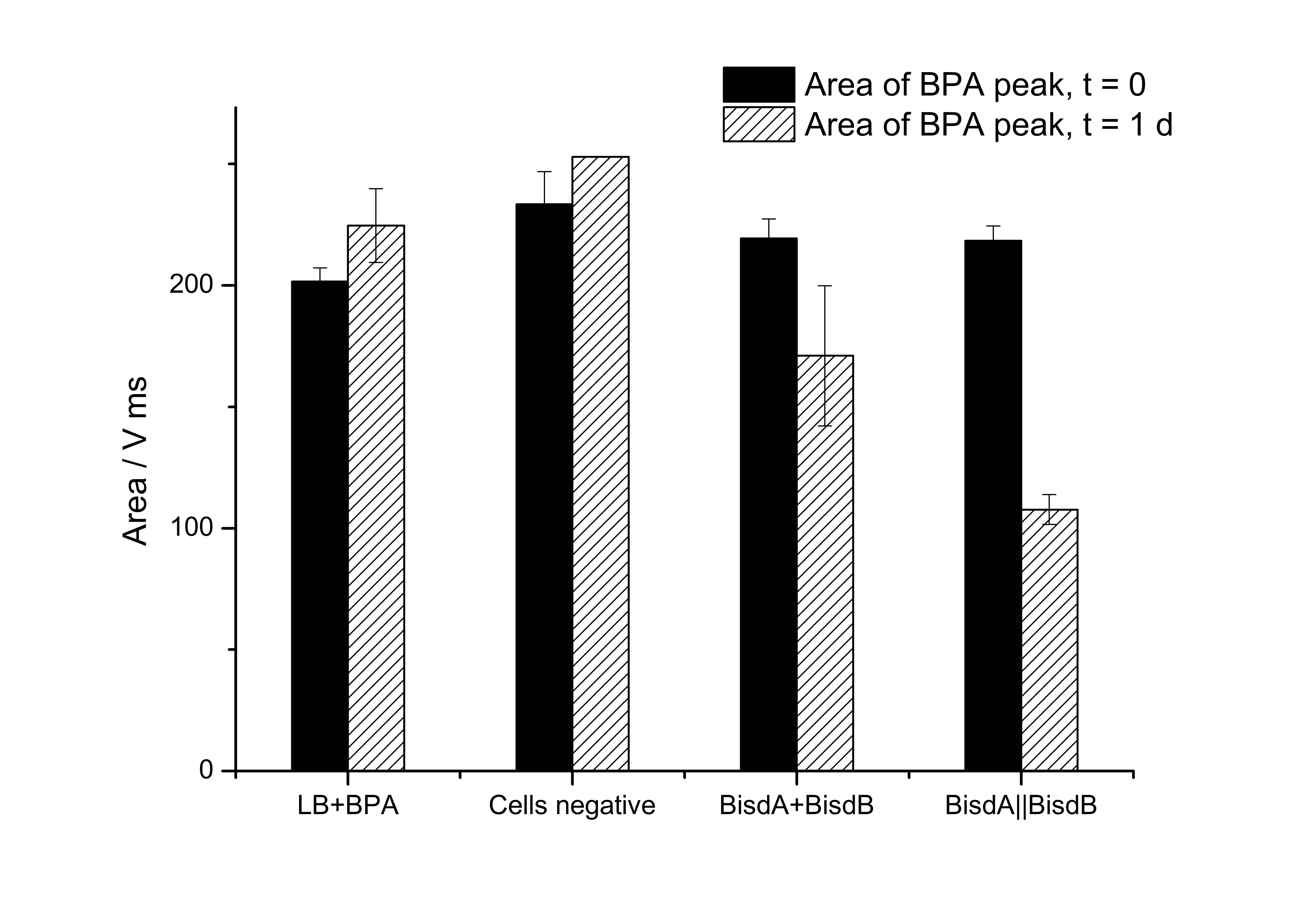
[[Image:Bielefeld2011_BPAAbbau_Versuch_1.jpg|300px|thumb|right| '''Figure 1: HPLC results of the first experiment on BPA degradation in ''E. coli'' TOP10. Cultivations were carried out in LB medium with 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein. ''']] | [[Image:Bielefeld2011_BPAAbbau_Versuch_1.jpg|300px|thumb|right| '''Figure 1: HPLC results of the first experiment on BPA degradation in ''E. coli'' TOP10. Cultivations were carried out in LB medium with 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken at the beginning of the cultivation and after one day. The HPLC results are shown above (area of the BPA peak). BisdA + BisdB is the polycistronic gene and BisdABisdB is the fusion protein. ''']] | ||
| - | |||
Bisphenol A: | Bisphenol A: | ||
* assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system) | * assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system) | ||
| - | * HPLC results: | + | * HPLC results: Fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare Figure 1) |
| - | + | ||
S-layer: | S-layer: | ||
| - | * expression of the S-layer gene without TAT-sequence of '' | + | * expression of the S-layer gene without TAT-sequence of ''C. glutamicum'' in ''E. coli'' [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results |
* successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 ''Corynebacterium halotolerans''] | * successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 ''Corynebacterium halotolerans''] | ||
| - | |||
Organizational: | Organizational: | ||
* moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec] | * moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec] | ||
| + | <br style="clear: both" /> | ||
| - | + | ==Week 3: 16th - 22nd May== | |
| - | ==Week 3: 16th - 22nd | + | |
| - | + | ||
Bisphenol A: | Bisphenol A: | ||
| - | * successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP oxidoreductase] gene from ''E. coli'' TOP10 | + | * successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP<sup>+</sup> oxidoreductase] gene from ''E. coli'' TOP10 |
| - | + | ||
S-layer: | S-layer: | ||
| - | * successful cloning of the complete S-layer gene ''cspB'' of '' | + | * successful cloning of the complete S-layer gene ''cspB'' of ''C. glutamicum'', ''C. crenatum'' and ''C. halotolerans'' |
* successful cloning of the S-layer genes of ''B. flavum'' and ''C. halotolerans'' without TAT-sequence, without lipid anchor and without both (only self-assembly domain) | * successful cloning of the S-layer genes of ''B. flavum'' and ''C. halotolerans'' without TAT-sequence, without lipid anchor and without both (only self-assembly domain) | ||
| - | + | ==Week 4: 23rd - 29th May== | |
| - | + | ||
| - | + | ||
| - | ==Week 4: 23rd - 29th | + | |
| - | + | ||
Bisphenol A: | Bisphenol A: | ||
* establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS) | * establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS) | ||
| - | |||
| - | |||
| - | |||
| - | |||
Organizational: | Organizational: | ||
| - | * arrange a BBQ for our workgroup in the CeBiTec to get to know our | + | * arrange a BBQ for our workgroup in the CeBiTec to get to know our co-workers |
* substantiating our contribution to the GENIALE | * substantiating our contribution to the GENIALE | ||
| - | + | ==Week 5: 30th May - 5th June== | |
| - | ==Week 5: 30th | + | |
| - | + | ||
Bisphenol A: | Bisphenol A: | ||
* beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>) | * beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>) | ||
| - | |||
| - | |||
| - | |||
| - | |||
Organizational: | Organizational: | ||
| - | * meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in | + | * meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in July |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Week 6: 6th - 12th June== | ||
Organizational: | Organizational: | ||
* presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention] | * presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention] | ||
| - | + | ==Week 7: 13th - 19th June== | |
| - | ==Week 7: 13th - 19th | + | S-layer: |
| - | S- | + | * successful fusion of modified ''cspB'' genes of [http://hamap.expasy.org/proteomes/CORGL.html ''C. glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] with a monomeric RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. |
| - | * successful fusion of modified cspB genes of [http://hamap.expasy.org/proteomes/CORGL.html '' | + | * ''C.glutamicum'': |
| - | * '' | + | ** K525131: fusion of ''cspB'' including lipid anchor and TAT-sequence <partinfo>K525121</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | ** K525131: fusion of cspB including lipid anchor and TAT-sequence | + | ** K525133: fusion of ''cspB'' including lipid anchor without TAT-sequence <partinfo>K525123</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | ** K525133: fusion of cspB including lipid anchor without TAT-sequence | + | |
| - | + | ||
* ''C. halotolerans'': | * ''C. halotolerans'': | ||
| - | ** K525233: fusion of cspB including lipid anchor without TAT-sequence | + | ** K525233: fusion of ''cspB'' including lipid anchor without TAT-sequence <partinfo>K525223</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | ** K525224: fusion of cspB including TAT-sequence without lipid anchor | + | ** K525224: fusion of ''cspB'' including TAT-sequence without lipid anchor <partinfo>K525224</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] |
| - | + | * First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD<sub>600</sub>) but in a higher fluorescence level. The variations of the induction time point has no significant influence on growth and the progression of fluorescence. | |
| - | * First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD<sub>600</sub>) but in a higher fluorescence level | + | |
[[Image:Bielefeld2011 Kult 19.06 OD600.png|450px|thumb|left| '''Figure 2: Growth curve of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] | [[Image:Bielefeld2011 Kult 19.06 OD600.png|450px|thumb|left| '''Figure 2: Growth curve of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] | ||
| - | [[Image:Bielefeld2011_Kult_1906_RFU.png|450px|thumb|right|'''Figure 3: Fluorescence | + | [[Image:Bielefeld2011_Kult_1906_RFU.png|450px|thumb|right|'''Figure 3: Fluorescence development of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] expressing K525133 during cultivation using different inducer (L-rhamnose) concentrations and varying the time point of induction. Cultivations were carried out in LB medium. Inducing time points and inducer concentration are named with following indices: '''(E1)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,1 %; '''(E2)''' induced early (cultivation time = 2,75 h) L-rhamnose end concentration = 0,2 %; '''(L1)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,1 %; '''(L2)''' induced late (cultivation time = 4,25 h) L-rhamnose end concentration = 0,2 %''']] |
| - | + | ||
Bisphenol A: | Bisphenol A: | ||
* cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites | * cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites | ||
| - | ==Week 8: 20th - 26th | + | ==Week 8: 20th - 26th June== |
S-layer: | S-layer: | ||
* successful cloning of K525232 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | * successful cloning of K525232 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | ||
| - | ** K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] without lipid anchor and TAT-sequence | + | ** K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] without lipid anchor and TAT-sequence <partinfo>K525222</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]. |
| - | + | ||
| - | + | ||
Organizational: | Organizational: | ||
* finishing our first press release | * finishing our first press release | ||
* all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived | * all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived | ||
| - | ==Week 9: 27th | + | ==Week 9: 27th June - 3rd July== |
| - | + | EXAMS ! | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==Week 10: 4th July - 10th July== | ||
Bisphenol A: | Bisphenol A: | ||
| - | * experiments on the influence of temperature, | + | * experiments on the influence of temperature, promoter strength and the characteristics of the fusion protein <partinfo>K123000</partinfo> || <partinfo>K123001</partinfo> on BPA degradation |
| - | + | S-layer: | |
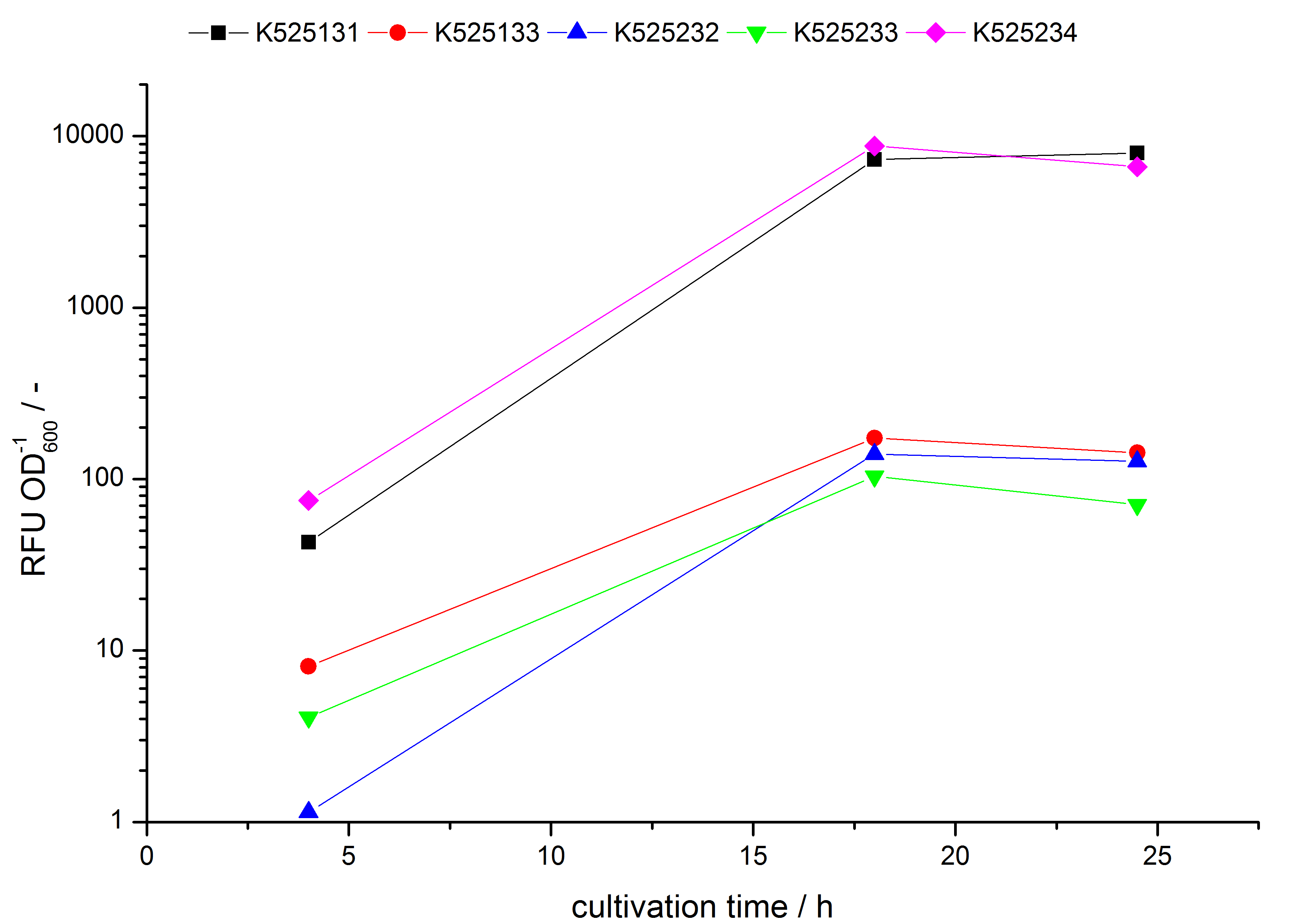
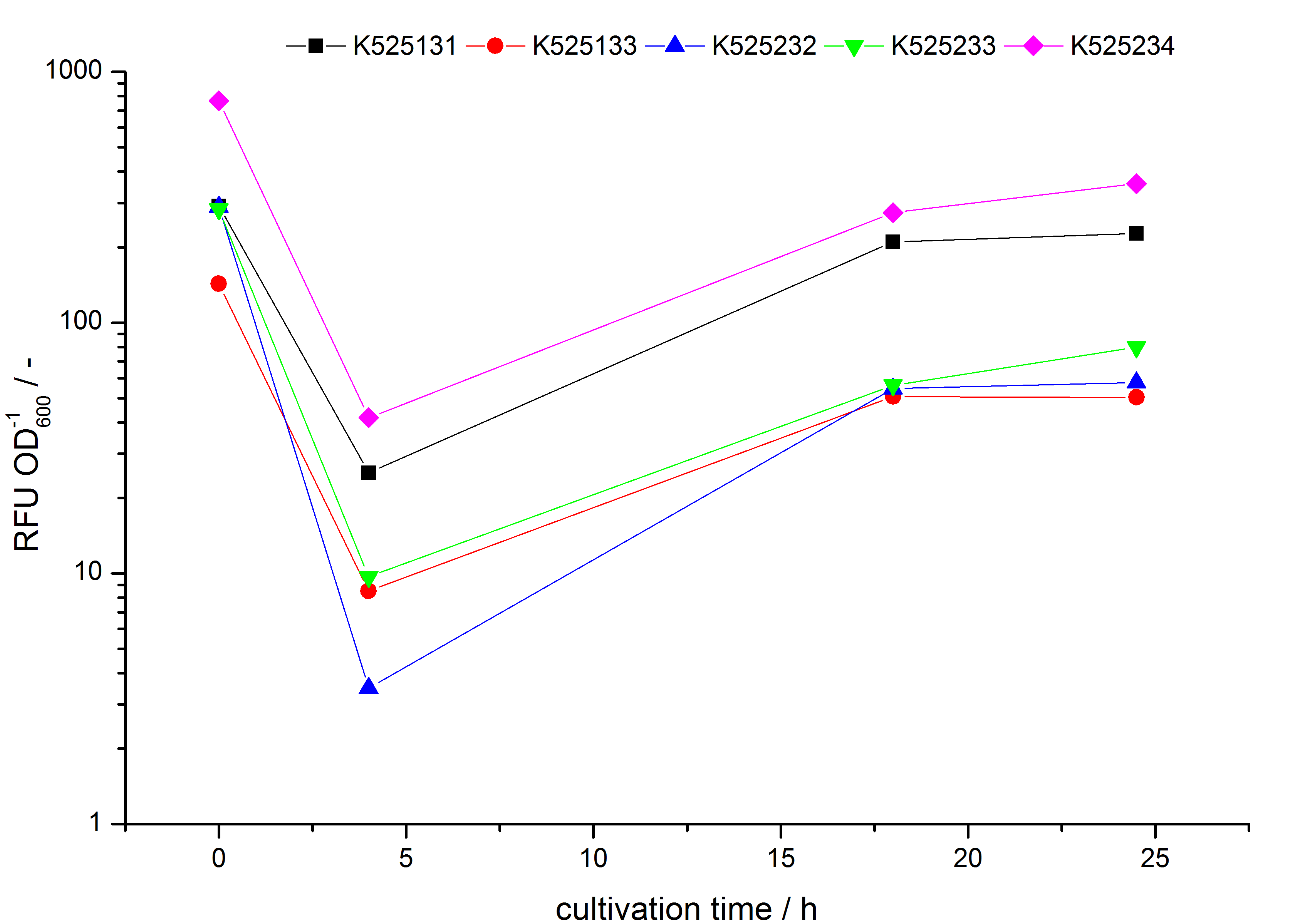
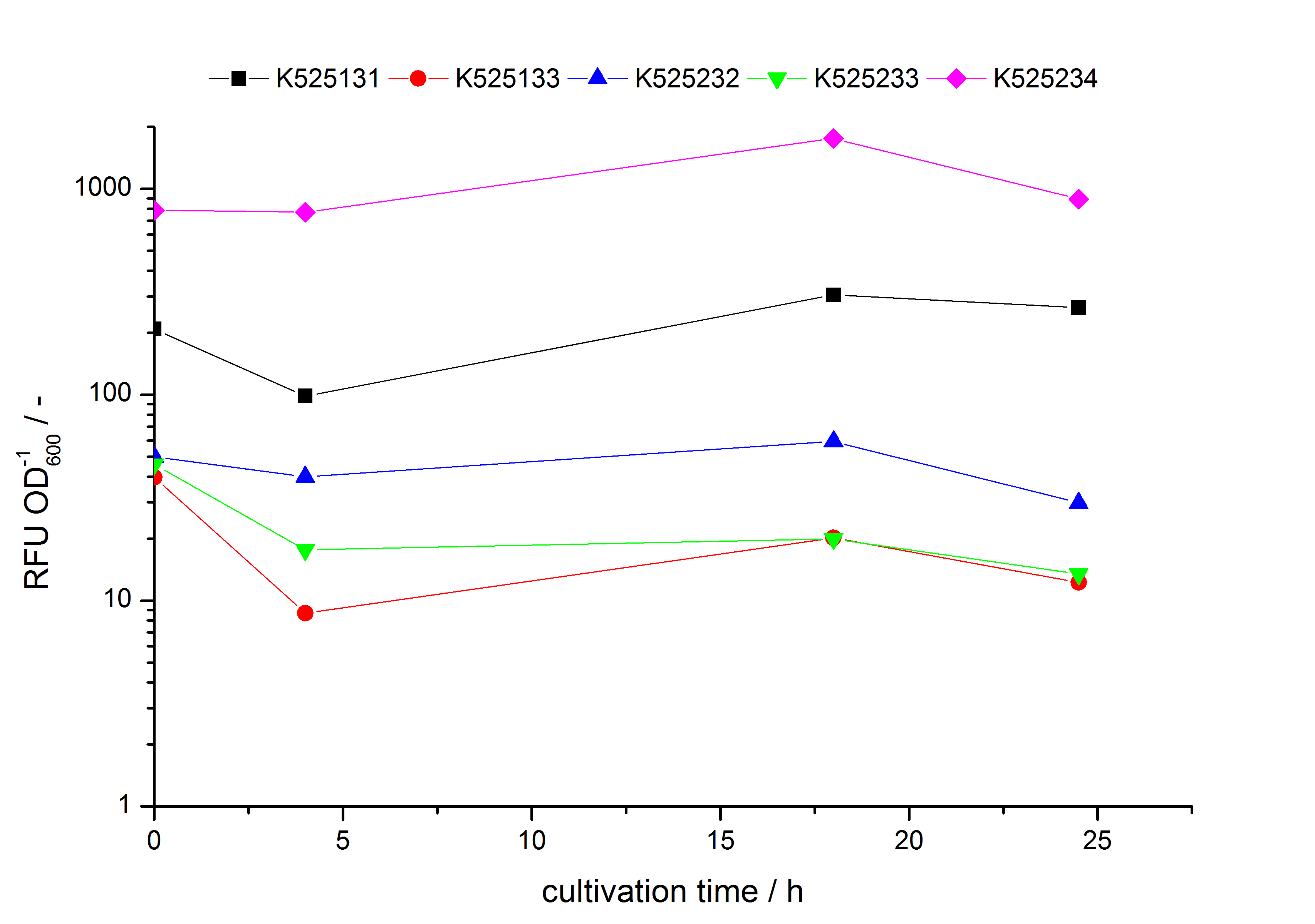
| - | S- | + | * Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in ''E. coli'' KRX to test the functional efficiency of ''Corynebacterium'' TAT-sequence in ''E.coli'' and the effect of the autoinduction protocol. After one day of cultivation, an increased fluorescence in all periplasm fractions of ''E. coli'' expressing S-layer constructs with TAT-sequence could be recognized. This is an indication that the ''Corynebacterium'' TAT-signal sequence is functional in ''E. coli''. Comparing the manual and the autoinduction protocol, no significant changes in fluorescence (after 14 h) are identifiable. |
| - | * Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in ''E. coli'' KRX to test the functional efficiency of Corynebacterium TAT-sequence in ''E.coli'' and the effect of the autoinduction protocol. After one day of cultivation | + | |
[[Image:Bielefeld2011 Kult 07.07 RFU pellet.png|450px|thumb|left| '''Figure 4: Relative fluorescence of the S-layer/mRFP fusion proteins in the cell suspension less the distinct fluorescence of ''E. coli'' KRX. All cultivations were carried out in LB medium at 37 ˚C except K525131<sub>Glc</sub> which was in autoinduction medium. The expression of the S-layers in LB medium were induced adding 0,1 % L-rhamnose at OD<sub>600</sub> = 0,6.''']] | [[Image:Bielefeld2011 Kult 07.07 RFU pellet.png|450px|thumb|left| '''Figure 4: Relative fluorescence of the S-layer/mRFP fusion proteins in the cell suspension less the distinct fluorescence of ''E. coli'' KRX. All cultivations were carried out in LB medium at 37 ˚C except K525131<sub>Glc</sub> which was in autoinduction medium. The expression of the S-layers in LB medium were induced adding 0,1 % L-rhamnose at OD<sub>600</sub> = 0,6.''']] | ||
| - | [[Image:Bielefeld2011 Kult 07.07 RFU periplasm.png|450px|thumb|right| '''Figure | + | [[Image:Bielefeld2011 Kult 07.07 RFU periplasm.png|450px|thumb|right| '''Figure 5: Relative fluorescence of the S-layer/mRFP fusion proteins in the periplasm less the distinct fluorescence of ''E. coli'' KRX periplasm. All cultivations were carried out in LB medium at 37 ˚C except K525131<sub>Glc</sub> which was in autoinduction medium. The expression of the S-layers in LB medium were induced adding 0,1 % L-rhamnose at OD<sub>600</sub> = 0,6.''']] |
| - | + | ||
Organizational: | Organizational: | ||
* presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research). | * presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research). | ||
| - | ==Week 11: 11th | + | ==Week 11: 11th July - 17th July== |
| - | + | Bisphenol A / S-layer: | |
| - | Bisphenol A / S- | + | * our BioBrick order (some fluorescent proteins and cleavage sites) from iGEM HQ arrived |
| - | * our BioBrick order (some | + | |
| - | + | ||
S-layer: | S-layer: | ||
| - | * our synthesized S- | + | * our synthesized S-layers SgsE and SbpA finally arrived |
| - | + | ||
| - | + | ||
Organizational: | Organizational: | ||
* presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos]) | * presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos]) | ||
| - | ==Week 12: 18th | + | ==Week 12: 18th July - 24th July== |
| - | + | ||
Organizational: | Organizational: | ||
* presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld | * presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld | ||
| - | * meeting | + | * meeting with the iGEM teams from Delft/NL, Edinburgh/UK, Odense/DK. Freiburg/DE and Ljubljana/SL at the Symposium ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627300144576/ Photos]) was fun! |
| - | + | ||
| - | + | ||
Bisphenol A / S-Layer: | Bisphenol A / S-Layer: | ||
| - | * removing illegal | + | * removing illegal restriction sites to get valid BioBricks |
| - | + | ||
| - | + | ||
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
| - | * cloning the NAD<sup>+</sup> dependent DNA ligase from ''E. coli'' into BioBrick backbones | + | * cloning the NAD<sup>+</sup>-dependent DNA ligase from ''E. coli'' into BioBrick backbones |
| - | ==Week 13: 25th | + | ==Week 13: 25th July - 31th July== |
| - | S- | + | S-layer: |
* fusing the synthesized S-Layers to a bunch of fluorescent proteins | * fusing the synthesized S-Layers to a bunch of fluorescent proteins | ||
* successful cloning of K525231 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | * successful cloning of K525231 using [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Gibson_assembly Gibson assembly]. | ||
| - | ** K525231: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] including lipid anchor and TAT-sequence | + | ** K525231: fusion of modified ''cspB'' of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] including lipid anchor and TAT-sequence <partinfo>K525221</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]. |
| - | * testing the basal transcription of ''E. coli'' KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD<sub>600</sub> reveals the basal transcription of ''E. coli'' KRX. L-rhamnose in the medium leads | + | * testing the basal transcription of ''E. coli'' KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD<sub>600</sub> reveals the basal transcription of ''E. coli'' KRX. L-rhamnose in the medium leads to a ten times higher RFU per OD<sub>600</sub>. |
| - | [[Image:Bielefeld2011 Kult 29.07 BF OD.png|450px|thumb|left| '''Figure 6: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html '' | + | [[Image:Bielefeld2011 Kult 29.07 BF OD.png|450px|thumb|left| '''Figure 6: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C. glutamicum'']. Cultivations were carried out in autoinduction medium at 37 ˚C with and without adding inducer (L-rhamnose). The induced cultivations are marked with index I and the uninduced are marked with index U.''']] |
[[Image:Bielefeld2011 Kult 29.07 CH OD.png|450px|thumb|right| '''Figure 7: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans'']. Cultivations were carried out in autoinduction medium at 37 ˚C with and without adding inducer (L-rhamnose). The induced cultivations are marked with index I and the uninduced are marked with index U.''']] | [[Image:Bielefeld2011 Kult 29.07 CH OD.png|450px|thumb|right| '''Figure 7: Growth curves of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans'']. Cultivations were carried out in autoinduction medium at 37 ˚C with and without adding inducer (L-rhamnose). The induced cultivations are marked with index I and the uninduced are marked with index U.''']] | ||
| - | |||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | [[Image:Bielefeld2011 Kult 29.07 RFUOD cells induced.png|450px|thumb|left| '''Figure 8: Development of relative fluorescence per OD<sub>600</sub> during cultivation of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html '' | + | [[Image:Bielefeld2011 Kult 29.07 RFUOD cells induced.png|450px|thumb|left| '''Figure 8: Development of relative fluorescence per OD<sub>600</sub> during cultivation of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C.glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans'']. Cultivations were carried out in autoinduction medium at 37 ˚C.''']] |
| - | [[Image:Bielefeld2011 Kult 29.07 RFUOD cells uninduced.png|450px|thumb|right| '''Figure 9: Development of relative fluorescence per OD<sub>600</sub> during uninduced cultivation of ''E. coli'' KRX containing fluorescence tagged S-layer gens constructs of [http://expasy.org/sprot/hamap/CORGL.html '' | + | [[Image:Bielefeld2011 Kult 29.07 RFUOD cells uninduced.png|450px|thumb|right| '''Figure 9: Development of relative fluorescence per OD<sub>600</sub> during uninduced cultivation of ''E. coli'' KRX containing fluorescence tagged S-layer gens constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C.glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans'']. Cultivations were carried out in autoinduction medium at 37 ˚C without adding inducer.''']] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Image:Bielefeld2011 Kult 29.07 RFUOD Periplasm.png|450px|thumb|centre| '''Figure 10: Development of relative fluorescence per OD<sub>600</sub> in periplasm of ''E. coli'' KRX expressing fluorescence tagged S-layer constructs of [http://expasy.org/sprot/hamap/CORGL.html ''C. glutamicum''] and [http://ijs.sgmjournals.org/content/54/3/779.abstract ''C. halotolerans''] during cultivation. Cultivations were carried out in autoinduction medium at 37 ˚C.''']] | ||
Bisphenol A: | Bisphenol A: | ||
* testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F) | * testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F) | ||
| - | ==Week 14: 1st | + | ==Week 14: 1st August - 7th August== |
Bisphenol A: | Bisphenol A: | ||
* BPA analysis with [[Team:Bielefeld-Germany/Protocols#Extraction_with_ethylacetate | extraction]] and LC-MS finally works and is very accurate | * BPA analysis with [[Team:Bielefeld-Germany/Protocols#Extraction_with_ethylacetate | extraction]] and LC-MS finally works and is very accurate | ||
* Better results for BPA degradation in ''E. coli'' -> our fusion protein (<partinfo>K123000</partinfo> to <partinfo>K123001</partinfo>) can completely degrade BPA | * Better results for BPA degradation in ''E. coli'' -> our fusion protein (<partinfo>K123000</partinfo> to <partinfo>K123001</partinfo>) can completely degrade BPA | ||
* Measuring characterization results for different BPA degrading BioBricks | * Measuring characterization results for different BPA degrading BioBricks | ||
| - | |||
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
| - | * Successful characterization of two differently labeled (6-FAM or TAMRA with Dabcyl) | + | * Successful characterization of two differently labeled (6-FAM or TAMRA with Dabcyl) molecular beacons as a preparation for the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#NAD.2B_bioassay NAD<sup>+</sup> bioassay] including autonomously produced NAD<sup>+</sup>-dependent DNA ligase (<partinfo>K525710</partinfo>) from ''E. coli'' (results shown below) |
| - | + | ||
| - | + | ||
| - | + | ||
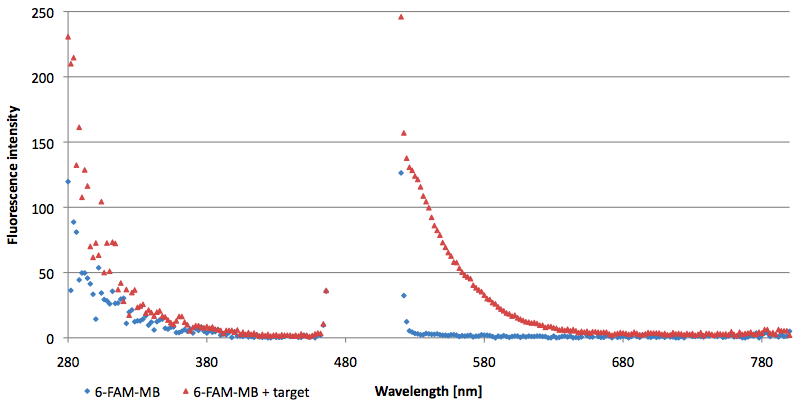
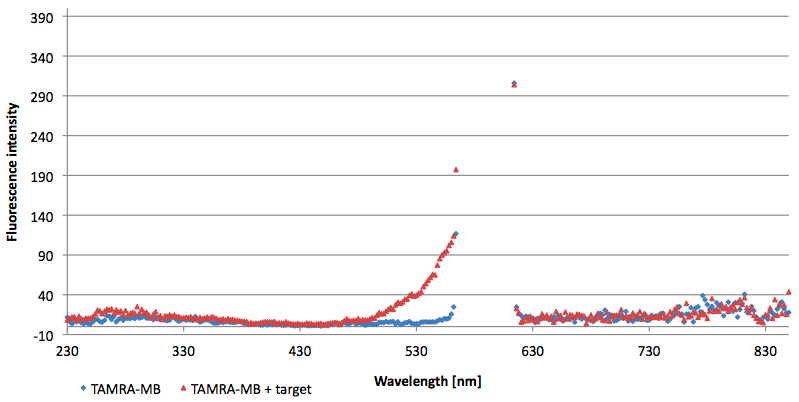
| + | [[Image:Bielefeld-Germany-2011 6FAM-emissionspectra-495nm target.png|450px|thumb|left| '''Figure 11: Emission spectra of 6-FAM labeled molecular beacon in its closed and open state (target added) at an extinction wavelength 495 nm (n=3).''']] | ||
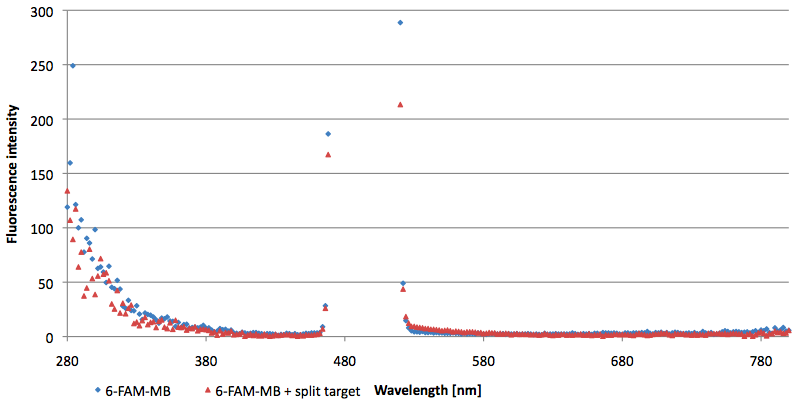
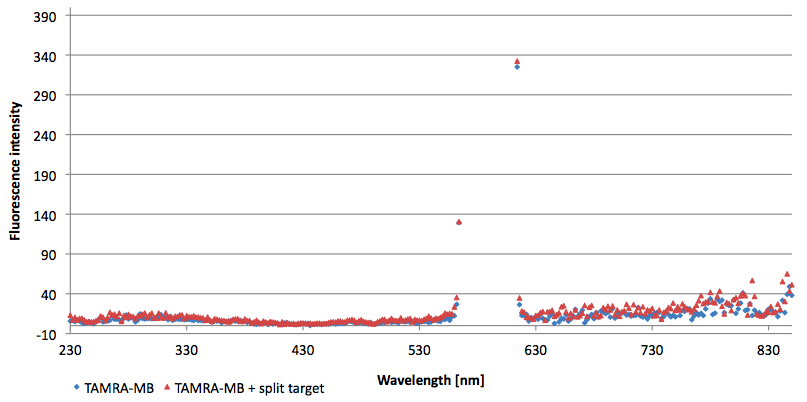
| + | [[Image:Bielefeld-Germany-2011 6FAM-emissionspectra-495nm split-target.png|450px|thumb|right| '''Figure 12: Emission spectra of 6-FAM labeled molecular beacon in its closed state (split target added) at an extinction wavelength 495 nm (n=3).''']] | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | [[Image:Bielefeld-Germany-2011 6FAM-extinctionspectra-530nm target.png|450px|thumb|left| '''Figure | + | [[Image:Bielefeld-Germany-2011 6FAM-extinctionspectra-530nm target.png|450px|thumb|left| '''Figure 13: Extinction spectra of 6-FAM labeled molecular beacon in its closed and open state (target added) at an emission wavelength 530 nm (n=3).''']] |
| - | [[Image:Bielefeld-Germany-2011 6FAM-extinctionspectra-530nm split-target.png|450px|thumb|right| '''Figure | + | [[Image:Bielefeld-Germany-2011 6FAM-extinctionspectra-530nm split-target.png|450px|thumb|right| '''Figure 14: Extinction spectra of 6-FAM labeled molecular beacon in its closed state (split target added) at an emission wavelength 530 nm (n=3).''']] |
| - | + | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
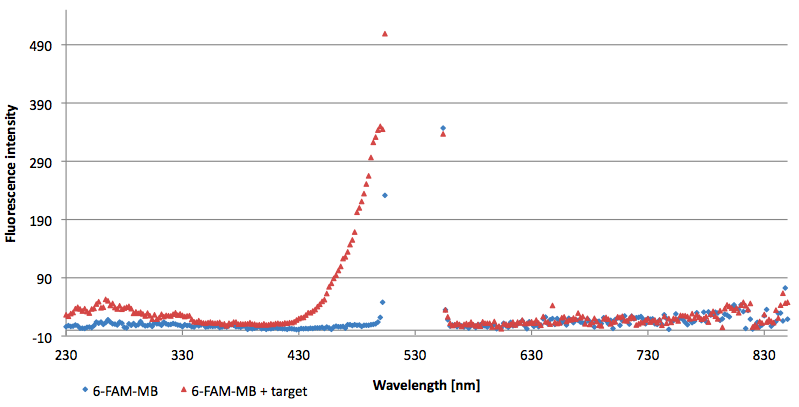
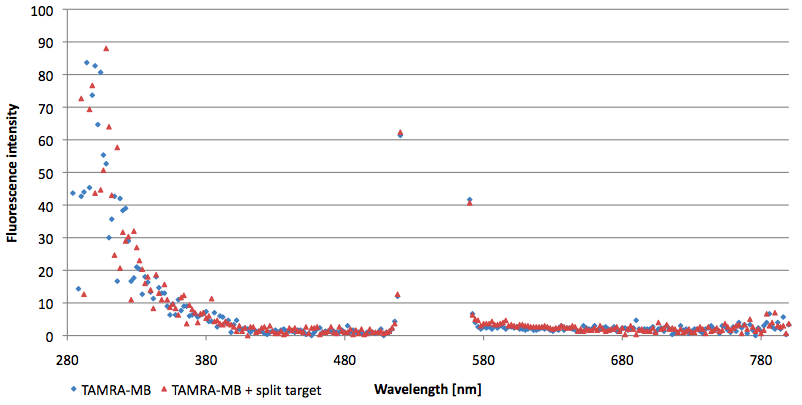
| - | [[Image:Bielefeld-Germany-2011 TAMRA-emissionspectra-544nm target.png|450px|thumb|left| '''Figure | + | [[Image:Bielefeld-Germany-2011 TAMRA-emissionspectra-544nm target.png|450px|thumb|left| '''Figure 15: Emission spectra of TAMRA labeled molecular beacon in its closed or open state (target added) at an extinction wavelength 544 nm (n=3).''']] |
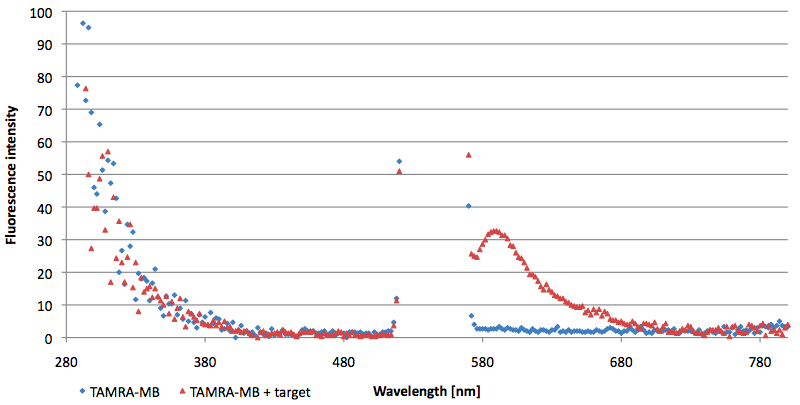
| - | [[Image:Bielefeld-Germany-2011 TAMRA-emissionspectra-544nm split-target.png|450px|thumb|right| '''Figure | + | [[Image:Bielefeld-Germany-2011 TAMRA-emissionspectra-544nm split-target.png|450px|thumb|right| '''Figure 16: Emission spectra of TAMRA labeled molecular beacon in its closed state (split target added) at an extinction wavelength 544 nm (n=3).''']] |
| - | + | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | [[Image:Bielefeld-Germany-2011 TAMRA-extinctionspectra-590nm target.png|450px|thumb|left| '''Figure | + | [[Image:Bielefeld-Germany-2011 TAMRA-extinctionspectra-590nm target.png|450px|thumb|left| '''Figure 17: Extinction spectra of TAMRA labeled molecular beacon in its closed or open state (target added) at an emission wavelength 590 nm (n=3).''']] |
| - | [[Image:Bielefeld-Germany-2011 TAMRA-extinctionspectra-590nm split-target.png|450px|thumb|right| '''Figure | + | [[Image:Bielefeld-Germany-2011 TAMRA-extinctionspectra-590nm split-target.png|450px|thumb|right| '''Figure 18: Extinction spectra of TAMRA labeled molecular beacon in its closed state (split target added) at an emission wavelength 590 nm (n=3).''']] |
| - | + | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
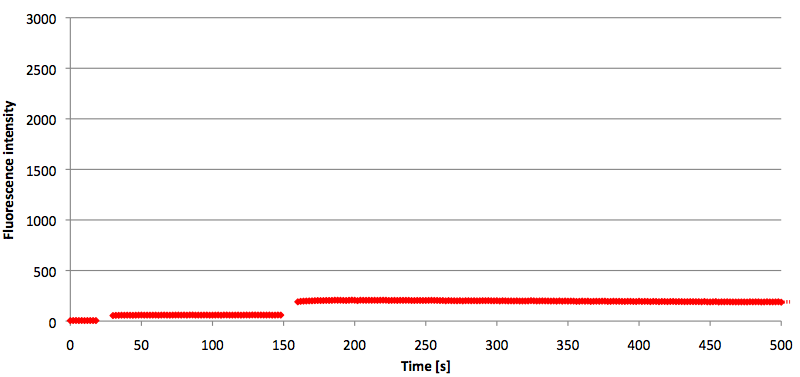
| - | [[Image:Bielefeld-Germany-2011 6FAM-signaltobackgroundratio-490-530nm target.png|450px|thumb|left| '''Figure | + | [[Image:Bielefeld-Germany-2011 6FAM-signaltobackgroundratio-490-530nm target.png|450px|thumb|left| '''Figure 19: Signal-to-background ratio (S/B) determination of 6-FAM labeled molecular beacon in its closed or open state at an extinction wavelength 495 nm and emission wavelength 530 nm. Molecular beacons and the target were added one after another (see gaps) each after equilibrium was reached. Calculated S/B: 45.52 (n=3).''']] |
| - | + | ||
| - | + | ||
| + | [[Image:Bielefeld-Germany-2011 6FAM-signaltobackgroundratio-490-530nm split-target.png|450px|thumb|right| '''Figure 20: Signal-to-background ratio (S/B) determination of 6-FAM labeled molecular beacon in its closed state at an extinction wavelength 495 nm and emission wavelength 530 nm. Molecular beacons and the split target were added one after another (see gaps) each after equilibrium was reached. Calculated S/B: 3.36 (n=3).''']] | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
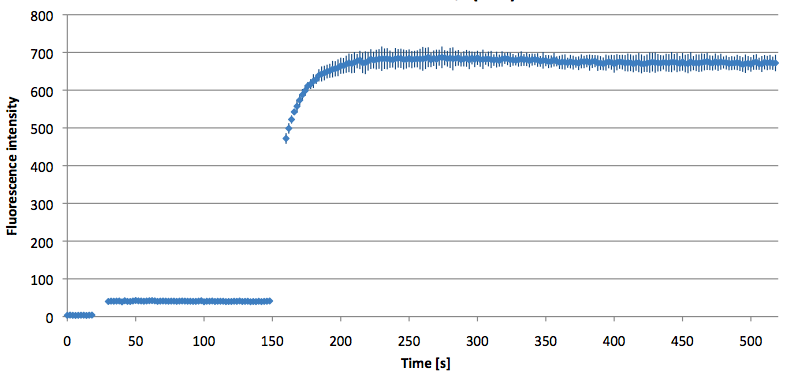
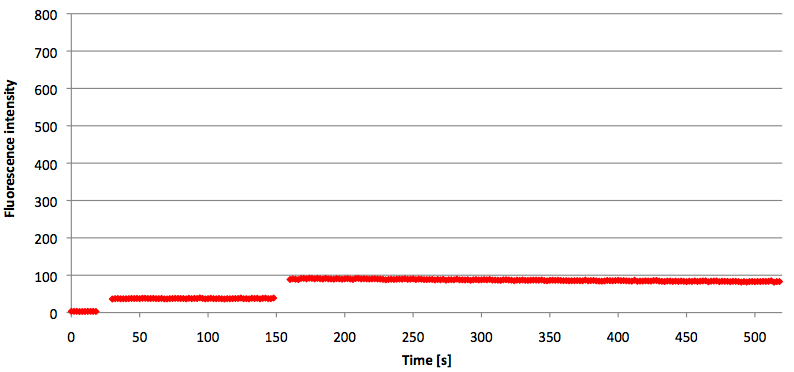
| - | [[Image:Bielefeld-Germany-2011 TAMRA-signaltobackgroundratio-552-590nm target.png|450px|thumb|left| '''Figure | + | [[Image:Bielefeld-Germany-2011 TAMRA-signaltobackgroundratio-552-590nm target.png|450px|thumb|left| '''Figure 21: Signal-to-background ratio (S/B) determination of TAMRA labeled molecular beacon in its closed or open state at an extinction wavelength 552 nm and emission wavelength 590 nm. Molecular beacons and the target were added one after another (see gaps) each after equilibrium was reached. Calculated S/B: 18.21 (n=3).''']] |
| - | + | ||
| - | + | ||
| + | [[Image:Bielefeld-Germany-2011 TAMRA-signaltobackgroundratio-552-590nm split-target.png|450px|thumb|right| '''Figure 22: Signal-to-background ratio (S/B) determination of TAMRA labeled molecular beacon in its closed state at an extinction wavelength 552 nm and emission wavelength 590 nm. Molecular beacons and the split target were added one after another (see gaps) each after equilibrium was reached. Calculated S/B: 2.31 (n=3).''']] | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
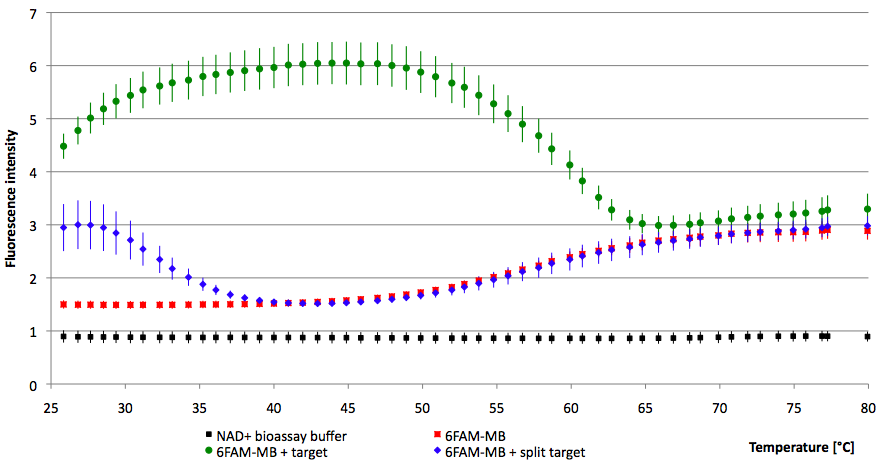
| - | [[Image:Bielefeld-Germany-2011 6FAM-thermal profile.png|450px|thumb|left| '''Figure | + | {| align="center" |
| - | + | || [[Image:Bielefeld-Germany-2011 6FAM-thermal profile.png|450px|thumb|left| '''Figure 23: Thermal profile of 6-FAM labeled molecular beacon alone and with either target or split target added (n=5).''']] | |
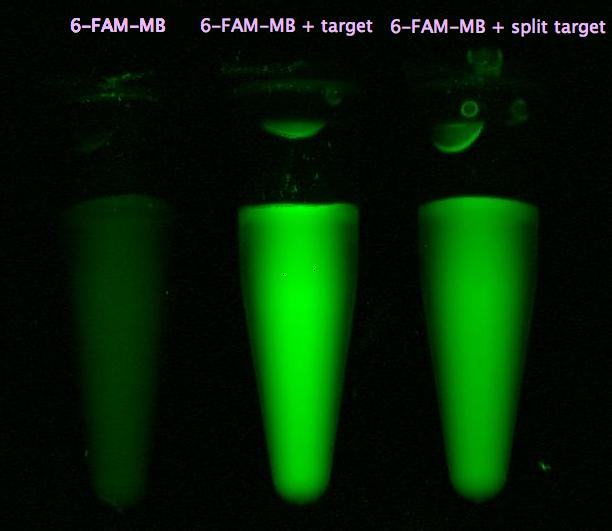
| - | + | || [[Image:Bielefeld-Germany-2011 6-FAM-MB image1.jpg|275px|thumb|right| '''Figure 24: Imaging 6-FAM labeled molecular beacon alone and with either target or split target added.''']] | |
| - | + | |} | |
| - | + | ||
| + | ==Week 15: 8th August - 14th August== | ||
Bisphenol A: | Bisphenol A: | ||
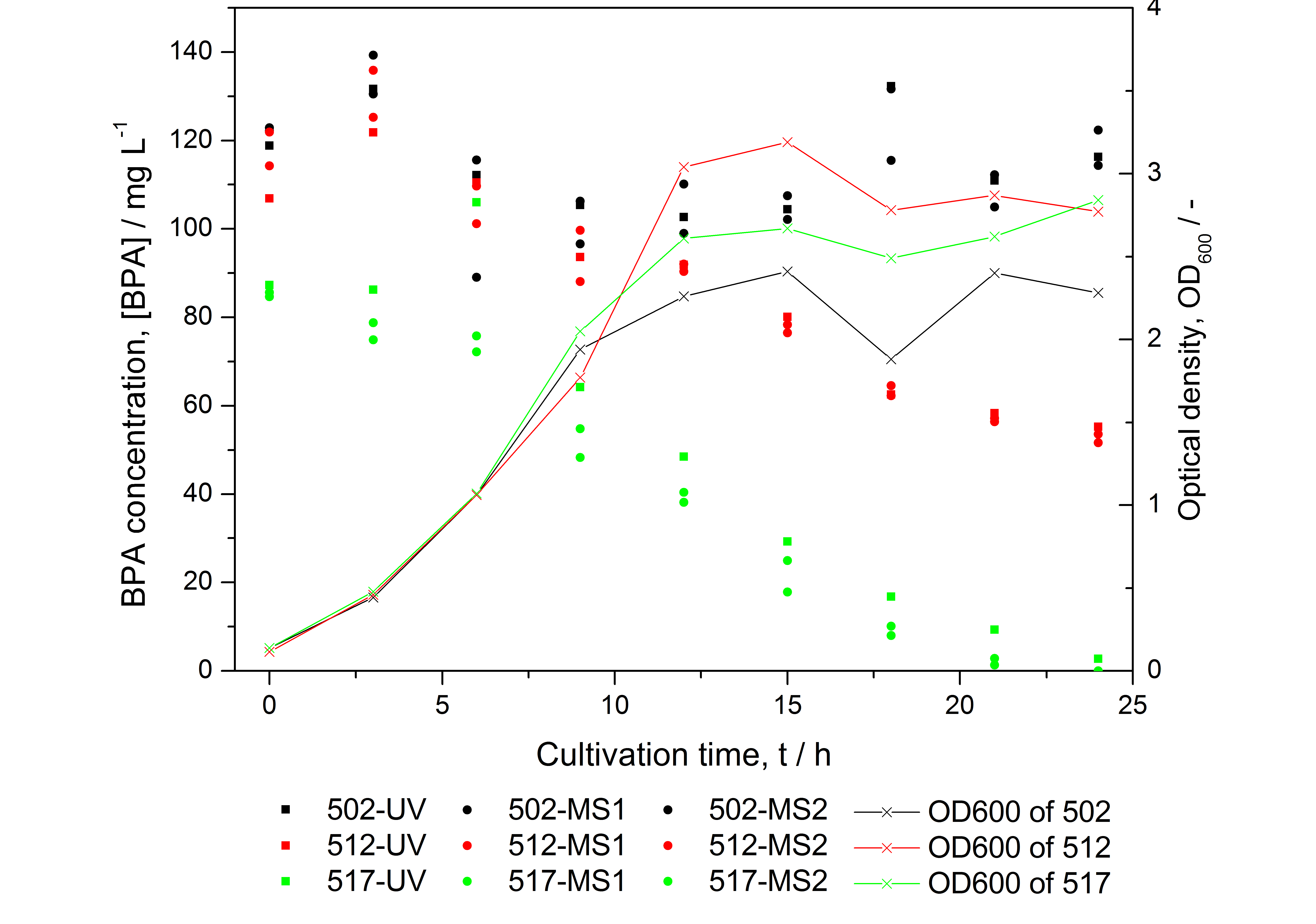
| - | * BPA analysis with [[Team:Bielefeld-Germany/Protocols#Extraction_with_ethylacetate | extraction]] and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare | + | * BPA analysis with [[Team:Bielefeld-Germany/Protocols/Analytics#Extraction_with_ethylacetate | extraction]] and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare Figure 25) |
* measuring of more samples from cultivations with BPA degrading BioBricks for further characterization | * measuring of more samples from cultivations with BPA degrading BioBricks for further characterization | ||
* we discovered some interesting results in our MS data - soon more | * we discovered some interesting results in our MS data - soon more | ||
* testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins | * testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins | ||
* testing the influence of BPA on the growth of ''E. coli'' | * testing the influence of BPA on the growth of ''E. coli'' | ||
| - | * developing a | + | * developing a model for BPA degradation by ''E. coli'' (compare Figure 26) |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Image:iGEM_Bielefeld_Vergleich_Analysemethoden_BPA.jpg|450px|thumb|left| '''Figure 25: HPLC results and optical density of experiments on BPA degradation of ''E. coli'' KRX. Cultivations were carried out in LB medium with ~ 100 mg L<sup>-1</sup> BPA at 30 °C. Samples were taken every three hours over one day. BPA concentration was measured by HPLC with either UV detector or ESI-qTOF-MS. 502: negative control (<partinfo>K123000</partinfo>), 512: polycistronic <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, 517: fusionprotein between <partinfo>K123001</partinfo> and <partinfo>K123000</partinfo>, every part behind medium strong constitutive promoter. ''']] | ||
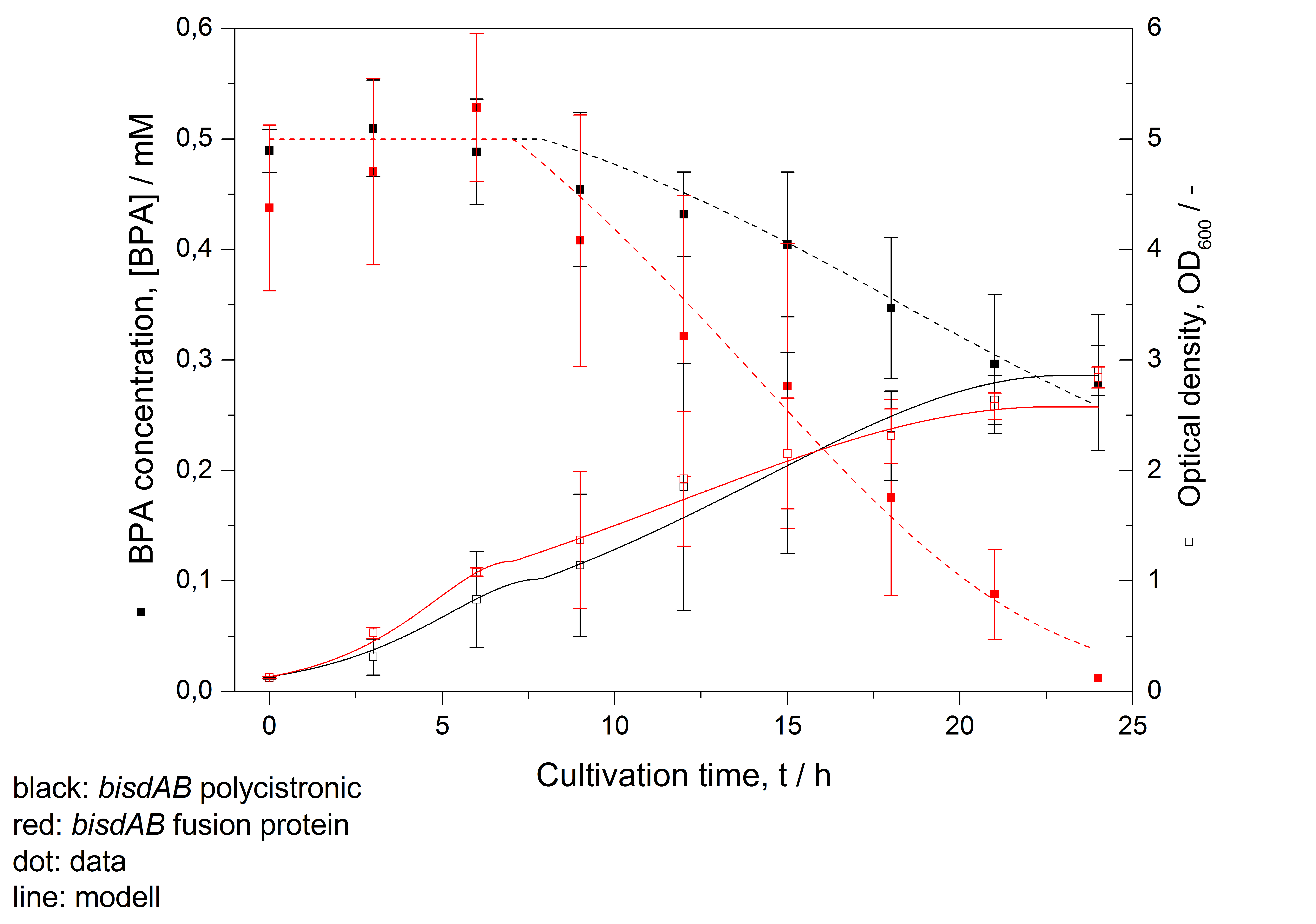
| + | [[Image:iGEM_Bielefeld2011_Verläufe_30°C_modelliert.jpg|450px|thumb|right| '''Figure 26: Modelling of BPA degradation (filled squares) by and OD<sub>600</sub> (open squares) of ''E. coli'' KRX carrying genes for BisdA and BisdB (polycistronic ''bisdAB'' (black) and fusion protein between BisdA and BisdB (red)) behind the medium strong promoter <partinfo>J23110</partinfo>. Cultivations were carried out at 30 °C in LB + Amp + BPA medium for 24 h with automatic sampling every three hours in 300 mL shaking flasks without baffles with silicon plugs. Three biological replicates were analysed. ''']] | ||
| + | <br style="clear: both" /> | ||
S-layer: | S-layer: | ||
| - | * MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by ''cspB'' gene) in ''E. coli'' | + | * MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by ''cspB'' gene) in ''E. coli'' KRX. |
| - | + | ||
| - | + | ||
| + | ==Week 16: 15th August - 21st August== | ||
S-layer: | S-layer: | ||
| - | * finally found our S-layer proteins in ''E. coli'' -> now we can plan purification strategy | + | * finally found our S-layer proteins in ''E. coli'' by analyzing the range of sizes in the polyacrylamid with a MALDI-TOF method-> now we can plan purification strategy |
* fusion of FPs to ''sgsE'' successful | * fusion of FPs to ''sgsE'' successful | ||
| - | |||
Bisphenol A: | Bisphenol A: | ||
* all metabolites of natural BPA degradation pathway found during degradation of BPA in ''E. coli'' by LC-MS -> now MS/MS to check structure of these metabolites to be sure | * all metabolites of natural BPA degradation pathway found during degradation of BPA in ''E. coli'' by LC-MS -> now MS/MS to check structure of these metabolites to be sure | ||
| Line 252: | Line 194: | ||
** seems like they can't | ** seems like they can't | ||
| - | ==Week 17: 22nd | + | ==Week 17: 22nd August - 28th August== |
| - | + | ||
S-layer: | S-layer: | ||
| - | * cultivation and purification of '' | + | * cultivation and test purification of ''SgsE'' fusion proteins (<partinfo>K525304</partinfo>, <partinfo>K525305</partinfo> and <partinfo>K525306</partinfo>) |
| - | + | ** SDS-PAGEs showed inclusion bodies formation in ''E coli'' KRX expressing ''sgsE''. | |
Bisphenol A: | Bisphenol A: | ||
* purification of his-tagged BisdA and BisdB | * purification of his-tagged BisdA and BisdB | ||
| Line 262: | Line 203: | ||
* cloning of fusion protein FNR:BisdA:BisdB successful | * cloning of fusion protein FNR:BisdA:BisdB successful | ||
| - | ==Week 18: 29th | + | ==Week 18: 29th August - 4th September== |
| + | S-layer: | ||
| + | * developing IEX clean-up for S-layers from ''Corynebacterium halotolerans'' | ||
| + | * Searching new purification methods for ''SgsE'' fusion proteins. | ||
| + | Bisphenol A: | ||
| + | * successful cloning of polycistronic FNR + BisdA + BisdB | ||
| + | NAD<sup>+</sup> detection: | ||
| + | *Successful overexpression of NAD<sup>+</sup>-dependent DNA ligase (<partinfo>K525710</partinfo>) in ''E. coli'' KRX and purification with Ni-NTA columns utilizing the protein`s C-terminal His-tag. Trying out utility for the NAD<sup>+</sup> bioassay now. | ||
| + | Organizational: | ||
| + | * participating at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld with a [http://www.geniale-bielefeld.de/programm/detail/date/456/show/Event/ lab tour] and a [http://www.geniale-bielefeld.de/programm/detail/date/295/show/Event/ presentation with public discussion afterwards (science café)]. Our participation is also mentioned in the regional newspaper [http://www.westfalen-blatt.de/nachricht/2011-08-30-wie-ein-hamster-im-laufrad/?cHash=b8421d55eb00ebf8532ebfa677a7e18f Westfalen Blatt] and in the internet on [http://www.direkt-bielefeld.de/Wissenschaft-im-Doppelpack-108856.html Bielefeld Direkt]. | ||
| + | ==Week 19: 5th September - 11th September== | ||
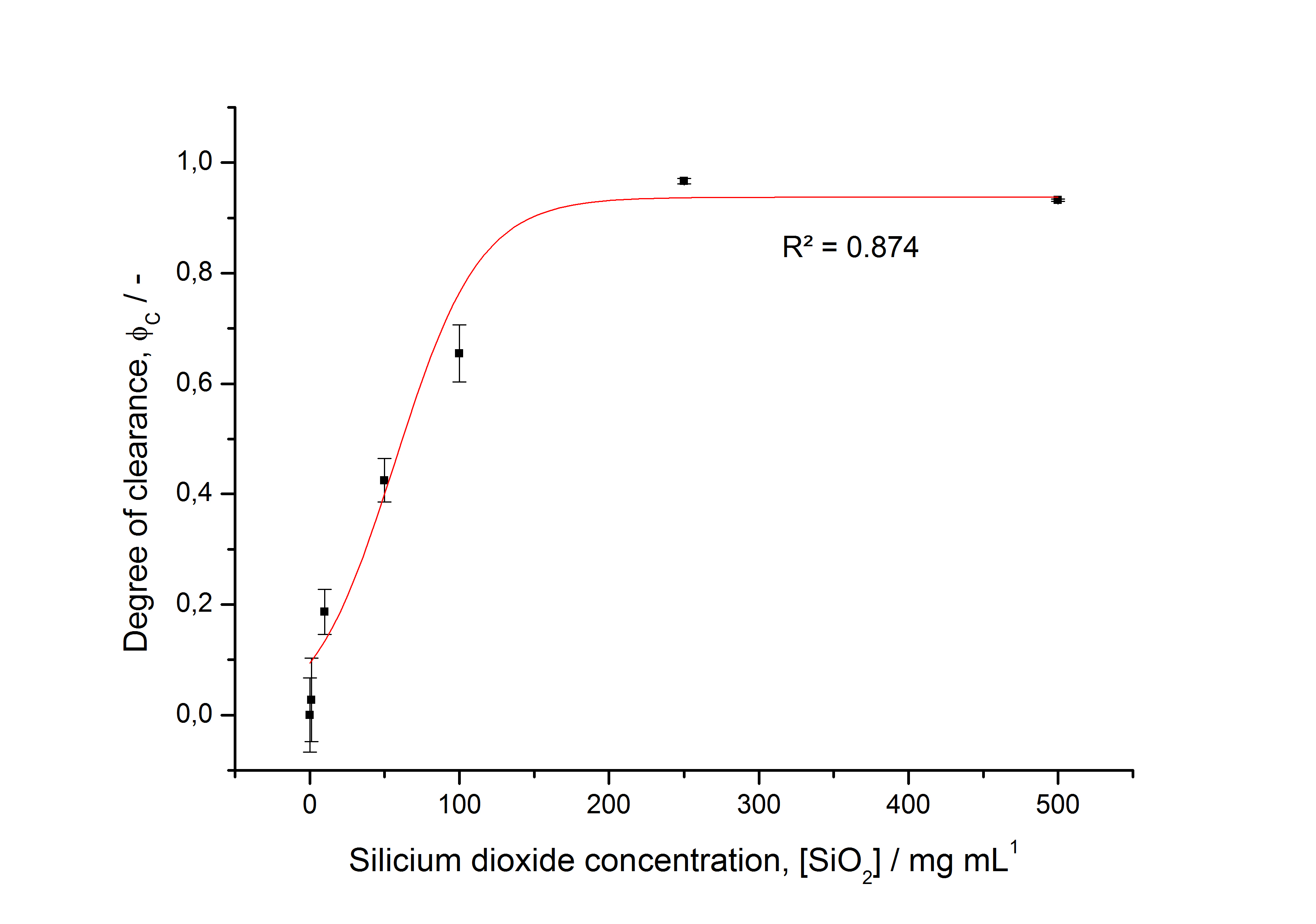
| + | [[Image:Bielefeld-Germany2011-saturationdegree305.jpg|500px|thumb|'''Fig. 27: Immobilization behaviour of fluorescent S-layer fusion protein SgsE :: mCitrine as a function of silica bead concentration. Data fitted with Michaelis-Menten function. ''']] | ||
S-layer: | S-layer: | ||
| + | * coating silica beads with fluorescent S-layer fusion protein SgsE | mCitrine, remove these proteins again and find them in SDS-PAGE | ||
| + | * characterizing immobilization behaviour of S-layers on silica beads -> seems to work (Figure 27) | ||
| + | * cultivate bigger amounts of and develop new purification strategy for S-layer fusion proteins | ||
| + | * testing purification of S-layer fusion proteins with his-tag | ||
| + | * cultivations of fusion protein between NADP<sup>+</sup>-ferredoxin oxidoreductase and SgsE / SbpA | ||
| + | * also SbpA | mCitrine fusion protein | ||
| + | NAD<sup>+</sup> detection: | ||
| + | * Suitability of autonomously produced DNA ligase (<partinfo>K525710</partinfo>) from ''E. coli'' for the NAD<sup>+</sup> bioassay could be demonstrated after treatment with NMN for deadenylation. Checking out the limit of detection right now. | ||
| + | <br style="clear: both" /> | ||
| + | ==Week 20: 12th September - 18th September== | ||
| + | S-layer: | ||
| + | * testing a purification and coating protocol for S-layer fusion protein mCitrine|SbpA. | ||
| + | * characterizing immobilization behaviour of mCitrine|SbpA [http://partsregistry.org/Part:BBa_K525405 (K525405)] on silica beads | ||
| + | * optimizing [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Production_and_purification_strategies purification] and [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Recrystallization_of_S-layer_proteins coating protocol] for ''SgsE'' fusion proteins. | ||
| + | * last characterization cultivations of [http://partsregistry.org/Part:BBa_K525304 K525304], [http://partsregistry.org/Part:BBa_K525305 K525305], [http://partsregistry.org/Part:BBa_K525306 K525306], [http://partsregistry.org/Part:BBa_K525405 K525405] and [http://partsregistry.org/Part:BBa_K525406 K525406]. | ||
| + | Bisphenol A: | ||
| + | * successful cloning of fusion protein FNR + BisdA | BisdB and polycistronic FNR + BisdA + BisdB behind constitutive promoter -> characterizing these last constructs | ||
| + | NAD<sup>+</sup> detection: | ||
| + | * NAD<sup>+</sup> assay works :) | ||
| - | + | ==Week 21: 19th September - 25th September== | |
| + | Organizational: | ||
| + | * lab work is finished, the last measurements and cloning experiments into pSB1C3 are done | ||
| + | * now it is time to fill the Wiki and Partsregistry with brilliant data until European Wikifreeze @ September 21st 11:59 pm EDT! | ||
| + | ==Week 22: 26th September - 2nd October== | ||
| + | Preparation of the presentation and the poster for the European Regional Jamboree. | ||
| + | * practicing the presentation with the Coryne and fermentation engineering working group as audience. | ||
| + | '''We are qualified for the final in Boston!!! Now labwork can continue.''' | ||
| + | |||
| + | ==Week 23: 3rd October - 9th October== | ||
| + | [[Image:Bielefeld-Germany-2011_Selectivity-of-LigA.png|450px|thumb|right| '''Figure 28: LigA shows high selectivity for NAD<sup>+</sup>.''' The final concentration of all analytes was 100 nM. The responses were evaluated on the basis of the average fluorescence enhancement rate in a range of 200 s after addition of each analyte into the NAD<sup>+</sup> bioassay. The dotted line marks the threshold indicating the intensity of background signal. All data are normalized to the NAD<sup>+</sup> value.]] | ||
| + | |||
| + | S-layer: | ||
| + | *scale-up cultivation of ''E. coli'' expressing [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] fusion protein. | ||
| + | *[https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#Fusion_proteins_of_SgsE_and_SbpA inclusion body purification] and testing further purification strategies. | ||
| + | *assembly of [http://partsregistry.org/Part:BBa_K525422 K525422] containing mCitrine|SbpA fused with a His-tag ([http://partsregistry.org/Part:BBa_K157011 K157011]) for easer purification. | ||
| + | *assembly of [http://partsregistry.org/Part:BBa_K525311 K525311] consisting of the SbpA S-layer fused with firefly-luciferase ([http://partsregistry.org/Part:BBa_K525999 K525999]) for testing an enzyme reaction on immobilized S-layer proteins. | ||
| + | *ordering silicon wafer as a new surface for immobilization. | ||
| + | *successful cloning of fusion protein of K525303 and K525560 [http://partsregistry.org/Part:BBa_K525001 (K525001)],[http://partsregistry.org/Part:BBa_K525311 (K525311)], [http://partsregistry.org/Part:BBa_K525006 SbpA|LigA (K525006)], [http://partsregistry.org/Part:BBa_K525422 (K525422)],[http://partsregistry.org/Part:BBa_K525322 (K525322)] | ||
NAD<sup>+</sup> detection: | NAD<sup>+</sup> detection: | ||
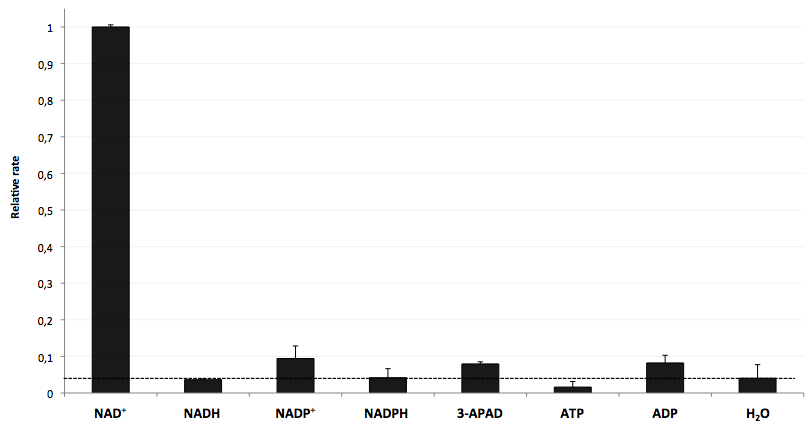
| - | * | + | * testing selectivity of LigA (<partinfo>BBa_K525710</partinfo>) for its substrate NAD<sup>+</sup> to verify further applications of the molecular beacon based NAD<sup>+</sup> bioassay dealing with NAD<sup>+</sup> detection in analyte mixtures such as cell lysates. |
| + | <br style="clear: both" /> | ||
| - | + | ==Week 24: 10th October - 16th October== | |
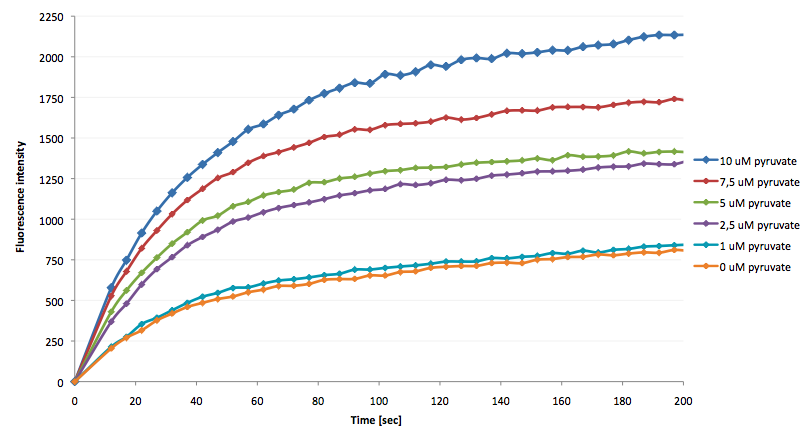
| - | * | + | [[Image:Bielefeld-Germany-2011 Fluorescence-enhancement-after addition-of-LDH-reaction-mix.png|450px|thumb|right| '''Figure 29: Fluorescence enhancement rate after addition of LDH reaction mix with various pyruvate concentrations into a LigA-dependent NAD<sup>+</sup> bioassay.''']] |
| + | |||
| + | S-layer: | ||
| + | *first expression of K525422 in ''E. coli'' KRX and His-tag affinity chromatography based purification using denaturing conditions. | ||
| + | **Test purifications were carried out using binding buffer with and without imidazole. | ||
| + | *new inclusion body purification of K525305 and K525405 to get new biological material to test the further purification methods. | ||
| + | *establishment of ion exchange chromatography (IEX) and hydrophobic interaction chromatography (HIC) as futher purification steps for S-layer proteins from ''Lysinibacillus sphaericus'' and ''Geobacillus stearothermophilus''. | ||
| + | **diethylaminoethyl cellulose (DEAE) as IEX and butyl sepharose™ 4 Fast Flow as HIC purification media. | ||
| + | *test and scale-up expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] in ''E. coli'' KRX. | ||
| + | **after enzymatic cell lysis with lysozyme and inclusion body purification highest luminescence was measured in the lysis supernatant. This implicates that the fusion protein does not form inclusion bodies. | ||
| + | *successful cloning of fusion protein of CspB and luciferase with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Genetics Gibson assembly] | ||
| + | NAD<sup>+</sup> detection: | ||
| + | *the NAD<sup>+</sup> bioassay was successfully coupled to the NADH-dependent reaction of lactic acid dehydrogenase and could therefore be used to quantify pyruvate | ||
| + | BPA-degradation: | ||
| + | *successful cloning of [http://partsregistry.org/Part:BBa_K525562 (K525562)] ( Fusion protein of [http://partsregistry.org/Part:BBa_K525499 (K525499)], [http://partsregistry.org/Part:BBa_K123000 (K123000)]and [http://partsregistry.org/Part:BBa_K123001 (K5123001)] with middle strong promoter) and [http://partsregistry.org/Part:BBa_K525534 (K525534)] | ||
| + | |||
| + | ==Week 25: 17th October - 23th October== | ||
| + | S-layer: | ||
| + | *new expression of <partinfo>K525311</partinfo> to develop an individual purification strategy without inclusion body purification. | ||
| + | *testing a new purification strategy for the fusion protein of the SgsE and the firefly-luciferase based on the IEX and HIC | ||
| + | *first expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] and successful His-tag affinity chromatography based purification using denaturing conditions. | ||
| + | NAD<sup>+</sup> detection: | ||
| + | *testing LigA for molecular cloning | ||
| + | |||
| + | ==Week 26: 24th October - 30th October== | ||
| + | S-layer: | ||
| + | *test expression of [http://partsregistry.org/Part:BBa_K525534 reductase|His-tag (K525534)] in ''E. coli'' KRX. and testing non denaturating His-tag affinity chromatography. | ||
| + | *recrystallization of [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] on silicon wafer using two different immobilization strategies. | ||
| + | *trying to get pictures of the S-layer nanostructures on coated silicon wafers using reflection electron microscope (REM) and atomic force microscopy (AFM) | ||
| + | **not enough time left to find a successful method | ||
Latest revision as of 23:09, 28 October 2011


Labjournal: On this page we summarize the (successful) results and achievements of our teamwork.
Week 1: 2nd - 8th May
Bisphenol A:
- cloning of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> behind weak (<partinfo>J23103</partinfo>) and medium strong (<partinfo>J23110</partinfo>) constitutive promoter (each part and both parts polycistronic)
- cloning of fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>, also assembly behind weak and medium strong constitutive promoter
- testing and establishing of HPLC method for [http://en.wikipedia.org/wiki/Bisphenol_A BPA] detection
- expression of the successfully assembled BPA degrading BioBricks in E. coli [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29 TOP10]
S-layer:
- successful PCR on the S-layer genes of [http://expasy.org/sprot/hamap/CORGL.html Corynebacterium glutamicum] and Corynebacterium crenatum
- successful cloning of the S-layer gene of Brevibacterium flavum without TAT-sequence
Organizational:
- meeting with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] to discuss our participation at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld
Week 2: 9th - 15th May
Bisphenol A:
- assembly of <partinfo>K157011</partinfo> behind existing BPA degrading parts (for purification and testing of <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> in a cell free system)
- HPLC results: Fusion protein between <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> can degrade BPA and seems to work better than the polycistronic version (compare Figure 1)
S-layer:
- expression of the S-layer gene without TAT-sequence of C. glutamicum in E. coli [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] and [http://openwetware.org/wiki/E._coli_genotypes#BL21.28DE3.29 BL21-Gold(DE3)] after sequencing gave correct results
- successful PCR on the S-layer gene of [http://ijs.sgmjournals.org/cgi/content/abstract/54/3/779 Corynebacterium halotolerans]
Organizational:
- moving to our own room in the [http://www.cebitec.uni-bielefeld.de/ CeBiTec]
Week 3: 16th - 22nd May
Bisphenol A:
- successful PCR on the [http://www.brenda-enzymes.org/php/result_flat.php4?ecno=1.18.1.2 NADP+ oxidoreductase] gene from E. coli TOP10
S-layer:
- successful cloning of the complete S-layer gene cspB of C. glutamicum, C. crenatum and C. halotolerans
- successful cloning of the S-layer genes of B. flavum and C. halotolerans without TAT-sequence, without lipid anchor and without both (only self-assembly domain)
Week 4: 23rd - 29th May
Bisphenol A:
- establishing a new method for analysis of BPA concentrations (extraction + LC-ESI-QTOF-MS)
Organizational:
- arrange a BBQ for our workgroup in the CeBiTec to get to know our co-workers
- substantiating our contribution to the GENIALE
Week 5: 30th May - 5th June
Bisphenol A:
- beginning of first characterization experiments for BPA degrading BioBricks (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>)
Organizational:
- meeting with Prof. [http://de.wikipedia.org/wiki/Alfred_Pühler Alfred Pühler] to plan our contribution to the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec symposium] in July
Week 6: 6th - 12th June
Organizational:
- presentation of the iGEM competition at the [http://www.bio.nrw.de/studentconvention 2nd BIO.NRW (PhD) Student Convention]
Week 7: 13th - 19th June
S-layer:
- successful fusion of modified cspB genes of [http://hamap.expasy.org/proteomes/CORGL.html C. glutamicum] and [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] with a monomeric RFP ([http://partsregistry.org/Part:BBa_E1010 BBa_E1010]) using Gibson assembly.
- C.glutamicum:
- K525131: fusion of cspB including lipid anchor and TAT-sequence <partinfo>K525121</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- K525133: fusion of cspB including lipid anchor without TAT-sequence <partinfo>K525123</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- C. halotolerans:
- K525233: fusion of cspB including lipid anchor without TAT-sequence <partinfo>K525223</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- K525224: fusion of cspB including TAT-sequence without lipid anchor <partinfo>K525224</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
- First expression of K525133 in [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX] to test different induction time points und L-rhamnose concentrations. A higher inducer concentration results in a decreasing maximum and final optical density (OD600) but in a higher fluorescence level. The variations of the induction time point has no significant influence on growth and the progression of fluorescence.
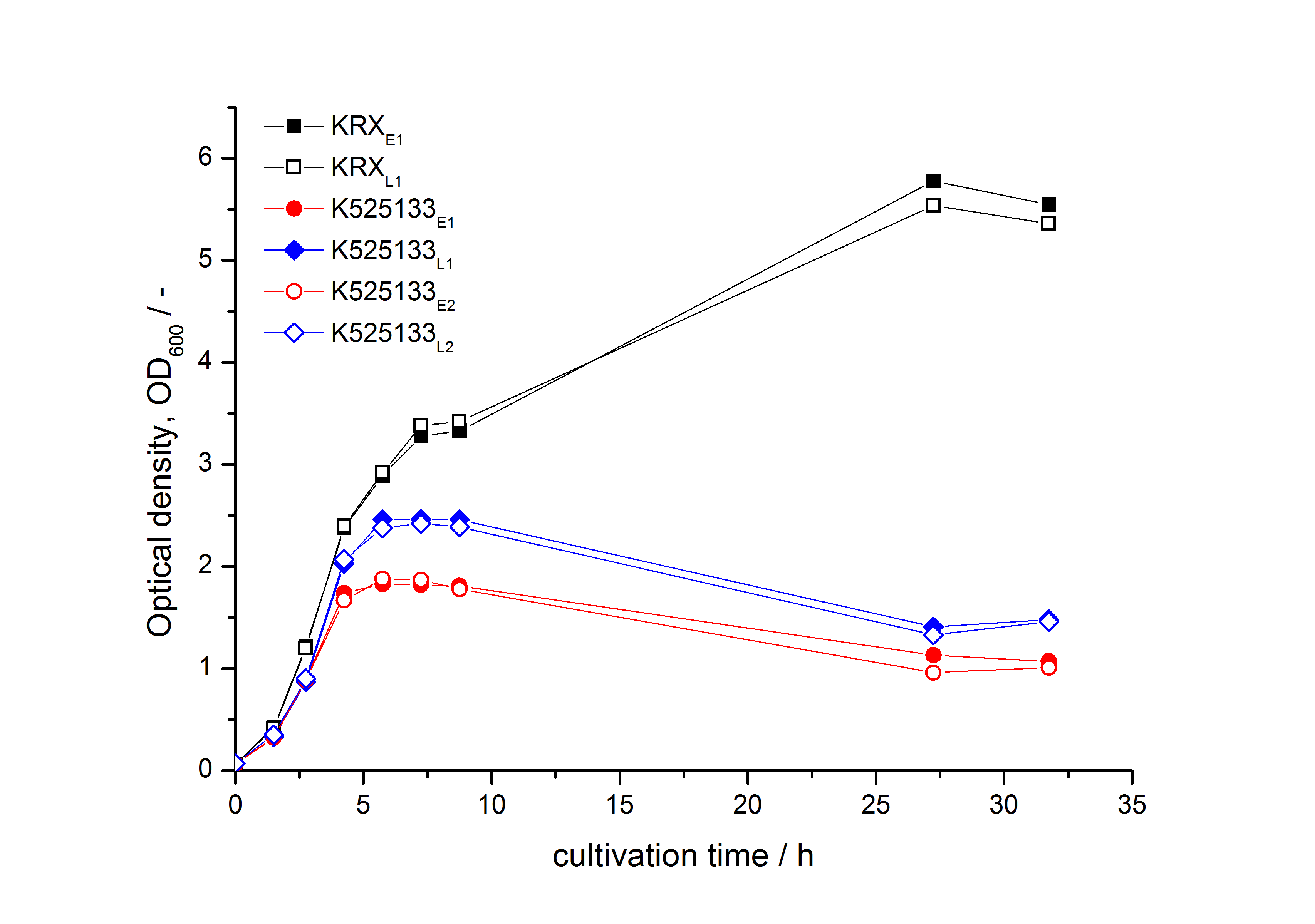
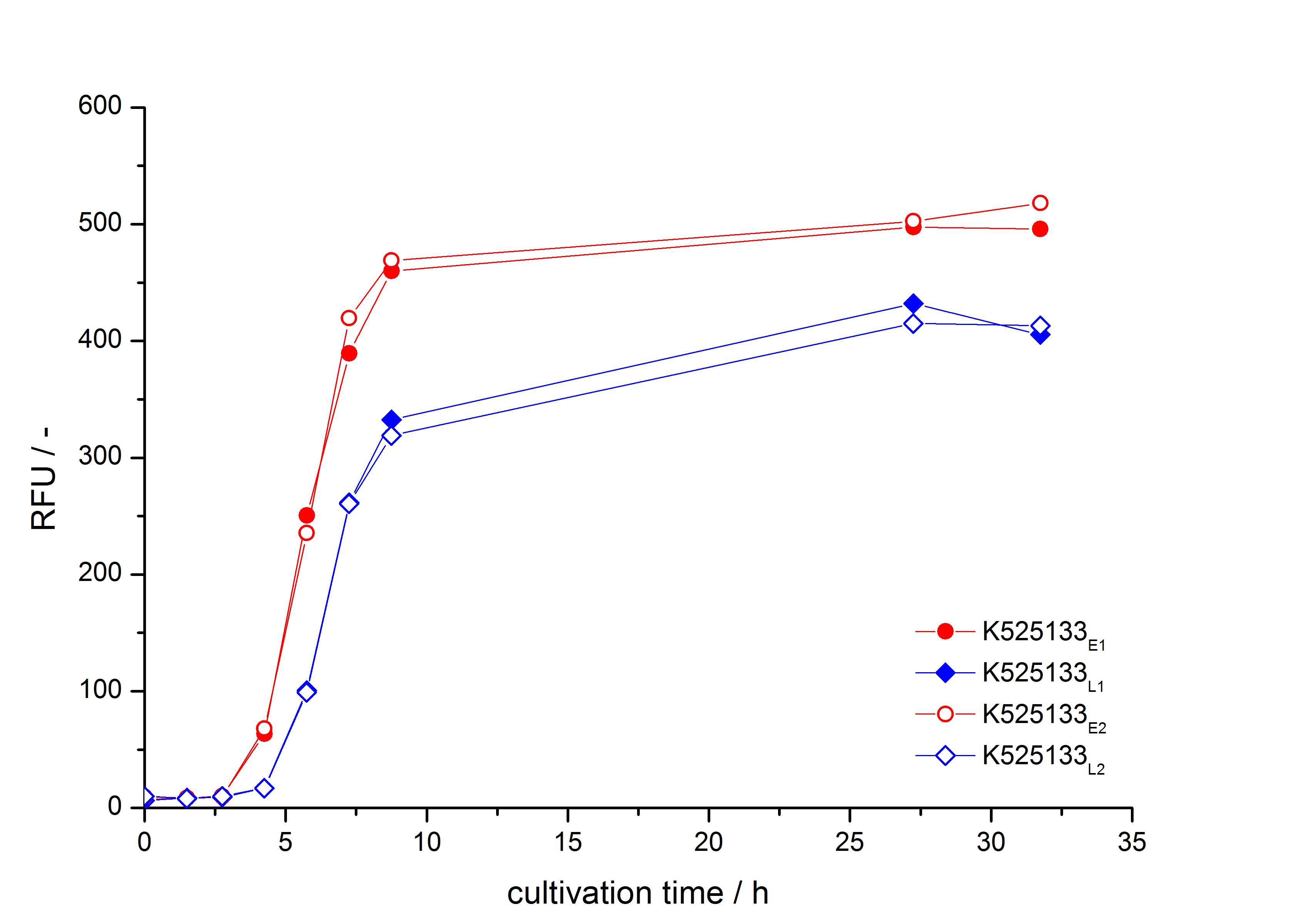
Bisphenol A:
- cloning of NADP oxidoreductase in [http://www.bioinfo.pte.hu/f2/pict_f2/pJETmap.pdf pJET1.2] finally successful -> waiting for sequencing results to remove illegal restriction sites
Week 8: 20th - 26th June
S-layer:
- successful cloning of K525232 using Gibson assembly.
- K525232: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] without lipid anchor and TAT-sequence <partinfo>K525222</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010].
Organizational:
- finishing our first press release
- all devices (thermocycler etc.) and materials (competent cells, polymerase, kits) from our sponsors arrived
Week 9: 27th June - 3rd July
EXAMS !
Week 10: 4th July - 10th July
Bisphenol A:
- experiments on the influence of temperature, promoter strength and the characteristics of the fusion protein <partinfo>K123000</partinfo> || <partinfo>K123001</partinfo> on BPA degradation
S-layer:
- Expression of K525131 (with TAT-sequence and lipid anchor), K5252133 (with lipid anchor), K525232 (nothing); K525233 (with lipid anchor) and K525234 (with TAT-sequence) in E. coli KRX to test the functional efficiency of Corynebacterium TAT-sequence in E.coli and the effect of the autoinduction protocol. After one day of cultivation, an increased fluorescence in all periplasm fractions of E. coli expressing S-layer constructs with TAT-sequence could be recognized. This is an indication that the Corynebacterium TAT-signal sequence is functional in E. coli. Comparing the manual and the autoinduction protocol, no significant changes in fluorescence (after 14 h) are identifiable.
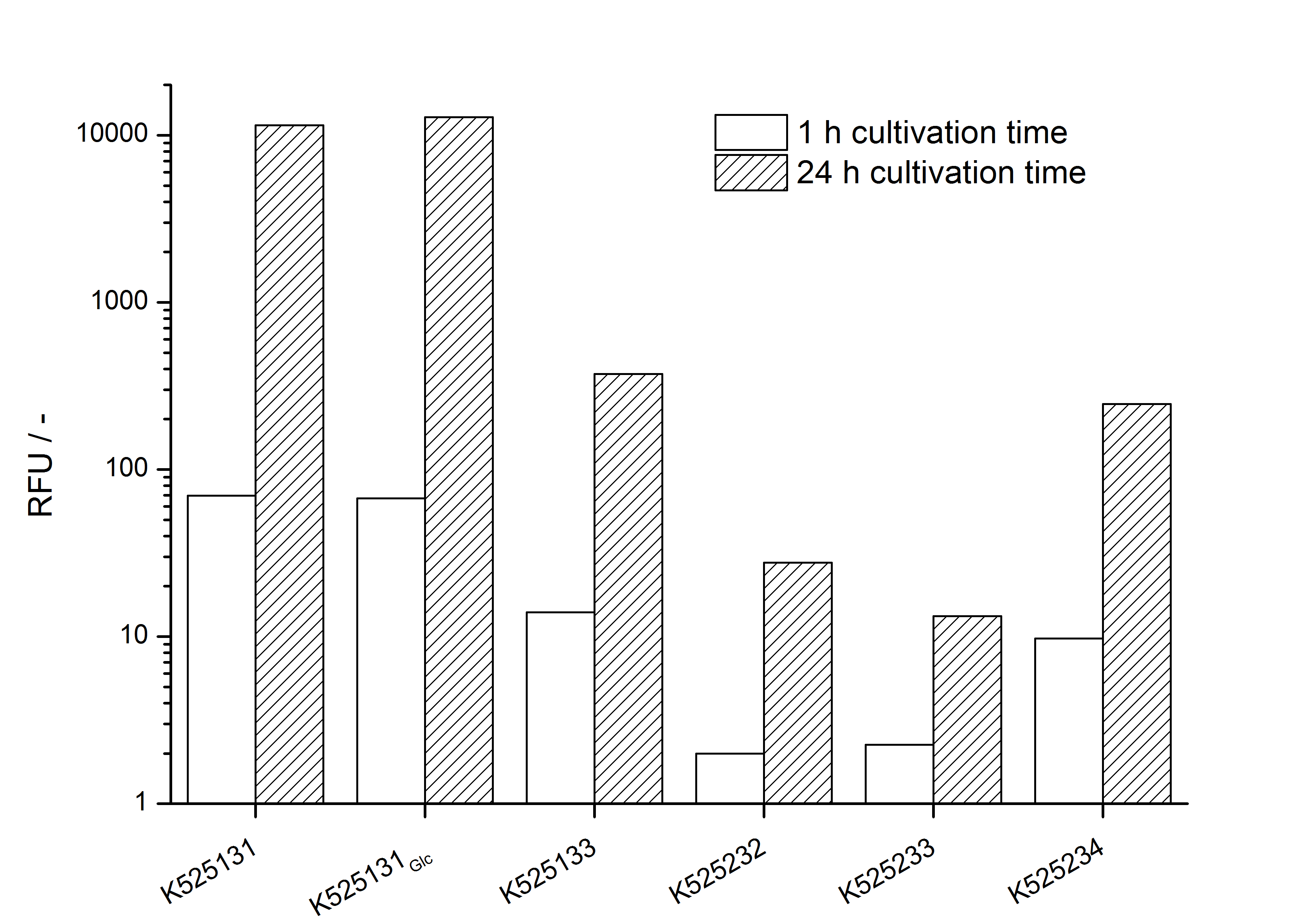

Organizational:
- presenting our posters from the teams of 2010 and 2011 at the congress [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=121262.html Biotechnologie2020+] in Berlin hosted by the "Bundesministerium für Bildung und Forschung" (Federal Ministry of Education and Research).
Week 11: 11th July - 17th July
Bisphenol A / S-layer:
- our BioBrick order (some fluorescent proteins and cleavage sites) from iGEM HQ arrived
S-layer:
- our synthesized S-layers SgsE and SbpA finally arrived
Organizational:
- presenting the iGEM competition and our team project at the secondary school [http://www.rg-herford.de/ Ravensberger Gynmasium] in Herford ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627214780020/ Photos])
Week 12: 18th July - 24th July
Organizational:
- presenting our projects from 2010 and 2011 at the [http://www.cebitec.uni-bielefeld.de/content/view/209/88/ CeBiTec Symposium 2011] in Bielefeld
- meeting with the iGEM teams from Delft/NL, Edinburgh/UK, Odense/DK. Freiburg/DE and Ljubljana/SL at the Symposium ([http://www.flickr.com/photos/igem-bielefeld/sets/72157627300144576/ Photos]) was fun!
Bisphenol A / S-Layer:
- removing illegal restriction sites to get valid BioBricks
NAD+ detection:
- cloning the NAD+-dependent DNA ligase from E. coli into BioBrick backbones
Week 13: 25th July - 31th July
S-layer:
- fusing the synthesized S-Layers to a bunch of fluorescent proteins
- successful cloning of K525231 using Gibson assembly.
- K525231: fusion of modified cspB of [http://ijs.sgmjournals.org/content/54/3/779.abstract C. halotolerans] including lipid anchor and TAT-sequence <partinfo>K525221</partinfo> with [http://partsregistry.org/Part:BBa_E1010 BBa_E1010].
- testing the basal transcription of E. coli KRX by adding inducer (L-rhamnose) or not. The increasing RFU per OD600 reveals the basal transcription of E. coli KRX. L-rhamnose in the medium leads to a ten times higher RFU per OD600.
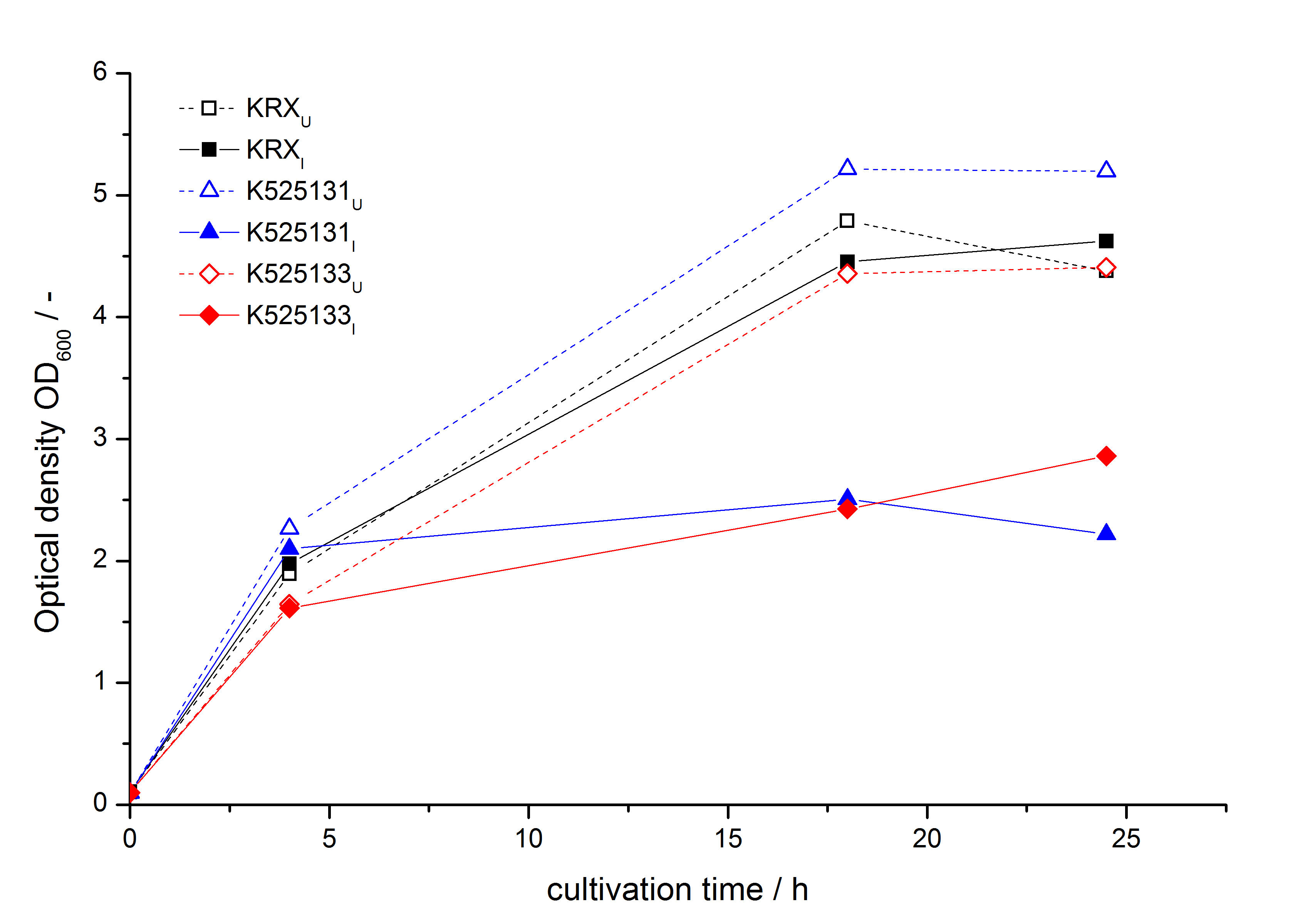

Bisphenol A:
- testing new BPA extraction protocols for LC-MS including an internal standard (bisphenol F)
Week 14: 1st August - 7th August
Bisphenol A:
- BPA analysis with extraction and LC-MS finally works and is very accurate
- Better results for BPA degradation in E. coli -> our fusion protein (<partinfo>K123000</partinfo> to <partinfo>K123001</partinfo>) can completely degrade BPA
- Measuring characterization results for different BPA degrading BioBricks
NAD+ detection:
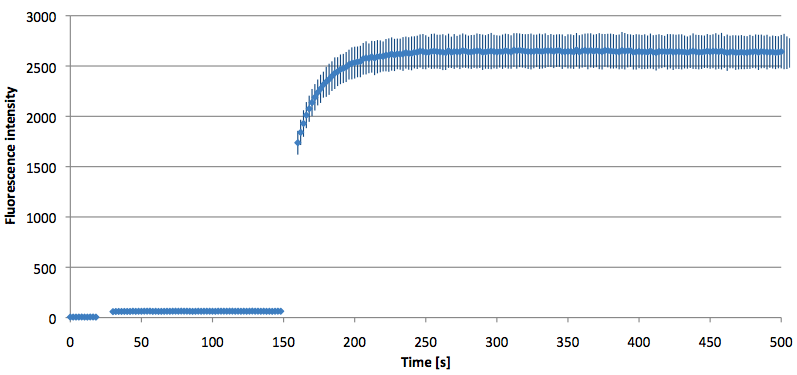
- Successful characterization of two differently labeled (6-FAM or TAMRA with Dabcyl) molecular beacons as a preparation for the NAD+ bioassay including autonomously produced NAD+-dependent DNA ligase (<partinfo>K525710</partinfo>) from E. coli (results shown below)
Week 15: 8th August - 14th August
Bisphenol A:
- BPA analysis with extraction and HPLC with UV detector leads to very similar results as the analysis with LC-MS (except for low BPA concentrations -> LOD / LOQ of LC-MS is lower than that of "normal" HPLC, compare Figure 25)
- measuring of more samples from cultivations with BPA degrading BioBricks for further characterization
- we discovered some interesting results in our MS data - soon more
- testing methods to purify his-tagged BisdA and BisdB for cell free BPA degradation and further characterization of these proteins
- testing the influence of BPA on the growth of E. coli
- developing a model for BPA degradation by E. coli (compare Figure 26)
S-layer:
- MALDI-TOF analysis of SDS-PAGEs to characterize the function of the TAT-sequence and the lipid anchor of PS2 (encoded by cspB gene) in E. coli KRX.
Week 16: 15th August - 21st August
S-layer:
- finally found our S-layer proteins in E. coli by analyzing the range of sizes in the polyacrylamid with a MALDI-TOF method-> now we can plan purification strategy
- fusion of FPs to sgsE successful
Bisphenol A:
- all metabolites of natural BPA degradation pathway found during degradation of BPA in E. coli by LC-MS -> now MS/MS to check structure of these metabolites to be sure
- some "metabolites" could also be found due to fragmentation during ionization of the product of BPA degradation
- testing whether E. coli can grow on BPA as the only carbon source (on M9 plates)
- seems like they can't
Week 17: 22nd August - 28th August
S-layer:
- cultivation and test purification of SgsE fusion proteins (<partinfo>K525304</partinfo>, <partinfo>K525305</partinfo> and <partinfo>K525306</partinfo>)
- SDS-PAGEs showed inclusion bodies formation in E coli KRX expressing sgsE.
Bisphenol A:
- purification of his-tagged BisdA and BisdB
- first characterization results entered into partsregistry
- cloning of fusion protein FNR:BisdA:BisdB successful
Week 18: 29th August - 4th September
S-layer:
- developing IEX clean-up for S-layers from Corynebacterium halotolerans
- Searching new purification methods for SgsE fusion proteins.
Bisphenol A:
- successful cloning of polycistronic FNR + BisdA + BisdB
NAD+ detection:
- Successful overexpression of NAD+-dependent DNA ligase (<partinfo>K525710</partinfo>) in E. coli KRX and purification with Ni-NTA columns utilizing the protein`s C-terminal His-tag. Trying out utility for the NAD+ bioassay now.
Organizational:
- participating at the science festival [http://www.geniale-bielefeld.de/ GENIALE] in Bielefeld with a [http://www.geniale-bielefeld.de/programm/detail/date/456/show/Event/ lab tour] and a [http://www.geniale-bielefeld.de/programm/detail/date/295/show/Event/ presentation with public discussion afterwards (science café)]. Our participation is also mentioned in the regional newspaper [http://www.westfalen-blatt.de/nachricht/2011-08-30-wie-ein-hamster-im-laufrad/?cHash=b8421d55eb00ebf8532ebfa677a7e18f Westfalen Blatt] and in the internet on [http://www.direkt-bielefeld.de/Wissenschaft-im-Doppelpack-108856.html Bielefeld Direkt].
Week 19: 5th September - 11th September
S-layer:
- coating silica beads with fluorescent S-layer fusion protein SgsE | mCitrine, remove these proteins again and find them in SDS-PAGE
- characterizing immobilization behaviour of S-layers on silica beads -> seems to work (Figure 27)
- cultivate bigger amounts of and develop new purification strategy for S-layer fusion proteins
- testing purification of S-layer fusion proteins with his-tag
- cultivations of fusion protein between NADP+-ferredoxin oxidoreductase and SgsE / SbpA
- also SbpA | mCitrine fusion protein
NAD+ detection:
- Suitability of autonomously produced DNA ligase (<partinfo>K525710</partinfo>) from E. coli for the NAD+ bioassay could be demonstrated after treatment with NMN for deadenylation. Checking out the limit of detection right now.
Week 20: 12th September - 18th September
S-layer:
- testing a purification and coating protocol for S-layer fusion protein mCitrine|SbpA.
- characterizing immobilization behaviour of mCitrine|SbpA [http://partsregistry.org/Part:BBa_K525405 (K525405)] on silica beads
- optimizing purification and coating protocol for SgsE fusion proteins.
- last characterization cultivations of [http://partsregistry.org/Part:BBa_K525304 K525304], [http://partsregistry.org/Part:BBa_K525305 K525305], [http://partsregistry.org/Part:BBa_K525306 K525306], [http://partsregistry.org/Part:BBa_K525405 K525405] and [http://partsregistry.org/Part:BBa_K525406 K525406].
Bisphenol A:
- successful cloning of fusion protein FNR + BisdA | BisdB and polycistronic FNR + BisdA + BisdB behind constitutive promoter -> characterizing these last constructs
NAD+ detection:
- NAD+ assay works :)
Week 21: 19th September - 25th September
Organizational:
- lab work is finished, the last measurements and cloning experiments into pSB1C3 are done
- now it is time to fill the Wiki and Partsregistry with brilliant data until European Wikifreeze @ September 21st 11:59 pm EDT!
Week 22: 26th September - 2nd October
Preparation of the presentation and the poster for the European Regional Jamboree.
- practicing the presentation with the Coryne and fermentation engineering working group as audience.
We are qualified for the final in Boston!!! Now labwork can continue.
Week 23: 3rd October - 9th October
S-layer:
- scale-up cultivation of E. coli expressing [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] fusion protein.
- inclusion body purification and testing further purification strategies.
- assembly of [http://partsregistry.org/Part:BBa_K525422 K525422] containing mCitrine|SbpA fused with a His-tag ([http://partsregistry.org/Part:BBa_K157011 K157011]) for easer purification.
- assembly of [http://partsregistry.org/Part:BBa_K525311 K525311] consisting of the SbpA S-layer fused with firefly-luciferase ([http://partsregistry.org/Part:BBa_K525999 K525999]) for testing an enzyme reaction on immobilized S-layer proteins.
- ordering silicon wafer as a new surface for immobilization.
- successful cloning of fusion protein of K525303 and K525560 [http://partsregistry.org/Part:BBa_K525001 (K525001)],[http://partsregistry.org/Part:BBa_K525311 (K525311)], [http://partsregistry.org/Part:BBa_K525006 SbpA|LigA (K525006)], [http://partsregistry.org/Part:BBa_K525422 (K525422)],[http://partsregistry.org/Part:BBa_K525322 (K525322)]
NAD+ detection:
- testing selectivity of LigA (<partinfo>BBa_K525710</partinfo>) for its substrate NAD+ to verify further applications of the molecular beacon based NAD+ bioassay dealing with NAD+ detection in analyte mixtures such as cell lysates.
Week 24: 10th October - 16th October
S-layer:
- first expression of K525422 in E. coli KRX and His-tag affinity chromatography based purification using denaturing conditions.
- Test purifications were carried out using binding buffer with and without imidazole.
- new inclusion body purification of K525305 and K525405 to get new biological material to test the further purification methods.
- establishment of ion exchange chromatography (IEX) and hydrophobic interaction chromatography (HIC) as futher purification steps for S-layer proteins from Lysinibacillus sphaericus and Geobacillus stearothermophilus.
- diethylaminoethyl cellulose (DEAE) as IEX and butyl sepharose™ 4 Fast Flow as HIC purification media.
- test and scale-up expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] in E. coli KRX.
- after enzymatic cell lysis with lysozyme and inclusion body purification highest luminescence was measured in the lysis supernatant. This implicates that the fusion protein does not form inclusion bodies.
- successful cloning of fusion protein of CspB and luciferase with Gibson assembly
NAD+ detection:
- the NAD+ bioassay was successfully coupled to the NADH-dependent reaction of lactic acid dehydrogenase and could therefore be used to quantify pyruvate
BPA-degradation:
- successful cloning of [http://partsregistry.org/Part:BBa_K525562 (K525562)] ( Fusion protein of [http://partsregistry.org/Part:BBa_K525499 (K525499)], [http://partsregistry.org/Part:BBa_K123000 (K123000)]and [http://partsregistry.org/Part:BBa_K123001 (K5123001)] with middle strong promoter) and [http://partsregistry.org/Part:BBa_K525534 (K525534)]
Week 25: 17th October - 23th October
S-layer:
- new expression of <partinfo>K525311</partinfo> to develop an individual purification strategy without inclusion body purification.
- testing a new purification strategy for the fusion protein of the SgsE and the firefly-luciferase based on the IEX and HIC
- first expression of [http://partsregistry.org/Part:BBa_K525322 mCitrine|SbpA|His-tag (K525322)] and successful His-tag affinity chromatography based purification using denaturing conditions.
NAD+ detection:
- testing LigA for molecular cloning
Week 26: 24th October - 30th October
S-layer:
- test expression of [http://partsregistry.org/Part:BBa_K525534 reductase|His-tag (K525534)] in E. coli KRX. and testing non denaturating His-tag affinity chromatography.
- recrystallization of [http://partsregistry.org/Part:BBa_K525305 mCitrine|SgsE (K525305)] and [http://partsregistry.org/Part:BBa_K525405 mCitrine|SbpA (K525405)] on silicon wafer using two different immobilization strategies.
- trying to get pictures of the S-layer nanostructures on coated silicon wafers using reflection electron microscope (REM) and atomic force microscopy (AFM)
- not enough time left to find a successful method
 "
"