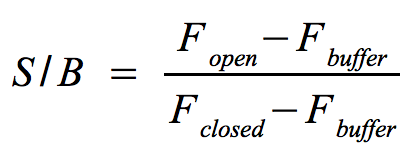Team:Bielefeld-Germany/Protocols/Analytics
From 2011.igem.org
(→Bradford Protein Assay) |
|||
| (42 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
==Bradford Protein Assay== | ==Bradford Protein Assay== | ||
| - | |||
For determination of protein concentrations in protein extracts from bacterial cell lysates or after protein purification steps a Bradford Protein Assay can be applied. For each sample three replicates and at least two dilutions should be measured. | For determination of protein concentrations in protein extracts from bacterial cell lysates or after protein purification steps a Bradford Protein Assay can be applied. For each sample three replicates and at least two dilutions should be measured. | ||
| - | + | *Prepare differentially concentrated BSA solutions in a range of 0-25 µg/mL for a BSA calibration curve. | |
| - | *Prepare differentially concentrated | + | |
*Fill a 96-well plate with 160 µL of each sample. | *Fill a 96-well plate with 160 µL of each sample. | ||
*Add 40 µL of Bradford [http://www.bio-rad.com/prd/en/US/adirect/biorad?cmd=catProductDetail&vertical=LSR&country=US&lang=en&productID=500-0205 Protein Assay Reagent] to each well and mix thoroughly by stirring as well as pipetting up and down. | *Add 40 µL of Bradford [http://www.bio-rad.com/prd/en/US/adirect/biorad?cmd=catProductDetail&vertical=LSR&country=US&lang=en&productID=500-0205 Protein Assay Reagent] to each well and mix thoroughly by stirring as well as pipetting up and down. | ||
| Line 14: | Line 12: | ||
*Use a plate reader for the measurement of absorbance at 595 nm. | *Use a plate reader for the measurement of absorbance at 595 nm. | ||
*Calculate the protein concentration with the help of the BSA calibration curve. | *Calculate the protein concentration with the help of the BSA calibration curve. | ||
| + | |||
==Fluorescence measurements== | ==Fluorescence measurements== | ||
| - | + | ===Measuring of [http://partsregistry.org/Part:BBa_E1010 mRFP] with Tecan Infinite® M200 platereader=== | |
| - | ===Measuring of [http://partsregistry.org/Part:BBa_E1010 mRFP]=== | + | |
* Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | * Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | ||
* Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | * Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | ||
| Line 30: | Line 28: | ||
** Integration time: 20 µs | ** Integration time: 20 µs | ||
| - | ===Measuring of [http://partsregistry.org/Part:BBa_J18931 mCitrine]=== | + | ===Measuring of [http://partsregistry.org/Part:BBa_J18931 mCitrine] with Tecan Infinite® M200 platereader=== |
* Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | * Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | ||
* Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | * Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | ||
| Line 39: | Line 37: | ||
** Excitation: 488 nm | ** Excitation: 488 nm | ||
** Emission: 529 nm | ** Emission: 529 nm | ||
| + | ** Number of reads: 25 | ||
| + | ** Manual gain: 75 | ||
| + | ** Integration time: 20 µs | ||
| + | |||
| + | ===Measuring of [http://partsregistry.org/Part:BBa_J18931 mCitrine] with Shimadzu RF-5301PC spectrofluorophotometer=== | ||
| + | * Fill 1 mL sample in a Plastibrand Disposable cuvette 1.5 mL semi-micro PS 12.5 x 12.5 x 45 mm by Brand | ||
| + | * Measure the fluorescence with following settings: | ||
| + | ** Exitation: 515 nm | ||
| + | ** Emission: 529 nm | ||
| + | ** 3 nm exitation filter | ||
| + | ** 10 nm emission filter | ||
| + | ** sensitivity high or low, depending on fluorescence in sample | ||
| + | |||
| + | |||
| + | ===Measuring of [http://partsregistry.org/Part:BBa_J18930 mCerulean] with Tecan Infinite® M200 platereader=== | ||
| + | * Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination | ||
| + | * Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark | ||
| + | * To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination) | ||
| + | * Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings: | ||
| + | ** 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm) | ||
| + | ** Measurement mode: Top | ||
| + | ** Excitation: 433 nm | ||
| + | ** Emission: 475 nm | ||
** Number of reads: 25 | ** Number of reads: 25 | ||
** Manual gain: 100 | ** Manual gain: 100 | ||
** Integration time: 20 µs | ** Integration time: 20 µs | ||
| - | |||
| + | == Luciferase Measurements == | ||
| + | For the luciferase detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer | ||
| + | |||
| + | |||
| + | For measuring cells: | ||
| + | * Prepare reaction tubes with 10 µL of high salt buffer (1M K<sub>2</sub>HPO<sub>4</sub>, 20mM EDTA, pH 7.8) | ||
| + | * Add 90 µL sample, mix and freeze at -80 °C | ||
| + | * For the measurement thaw by placing the tubes in room temperature water | ||
| + | * Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL<sup>-1</sup> lysozyme, 2.5 mg mL<sup>-1</sup> BSA, add water for desired volume) | ||
| + | * Mix and incubate the cells for 10 minutes at room temperature | ||
| + | |||
| + | For measuring cell-free samples start at this point: | ||
| + | * Prepare the Luciferase Assay Reagent, by adding 10 mL of Luciferase Assay Buffer to the vial containing the Luciferase Assay Substrate | ||
| + | * Fill each well of a white, flat bottom 96 well microtiter plate with 20 µL of cell lysate | ||
| + | * For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a [http://www.promega.com/b/de/Glomax/glomax_multi_plus.aspx Promega GloMax®-Multi+] Detection System with dual injector): | ||
| + | ** Injection volume of Luciferase Assay Reagent: 100 µL | ||
| + | ** Delay: 20 secs | ||
| + | ** Integration: 3 secs | ||
| + | |||
| + | ==Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)== | ||
This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility. | This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility. | ||
===Pouring the polyacrylamide gel=== | ===Pouring the polyacrylamide gel=== | ||
| - | + | *Make a master mix for the [[Team:Bielefeld-Germany/Protocols#SDS-PAGE_gel|stacking]] and [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#SDS-PAGE_gel separating gel] without adding ammonium persulfate and TEMED. | |
| - | *Make a master mix for the [ | + | |
*Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel. | *Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel. | ||
*Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into your gel casting form. Leave about 2 centimeters below the bottom of the comb for the stacking gel. | *Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into your gel casting form. Leave about 2 centimeters below the bottom of the comb for the stacking gel. | ||
| Line 63: | Line 102: | ||
===Preparing the sample=== | ===Preparing the sample=== | ||
| - | |||
*Mix your protein mixture 4:1 with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#4x_Laemmli-buffer Laemmli-buffer] (30 mL protein solution + 10 mL Laemmli-buffer) | *Mix your protein mixture 4:1 with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#4x_Laemmli-buffer Laemmli-buffer] (30 mL protein solution + 10 mL Laemmli-buffer) | ||
*Heat for 5 minutes at 95 °C. | *Heat for 5 minutes at 95 °C. | ||
===Running the gel=== | ===Running the gel=== | ||
| - | |||
*Remove sealing, put the polymerized gel into gel box and pour [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#SDS_running_buffer SDS running buffer] into the negative and positive electrode chamber. | *Remove sealing, put the polymerized gel into gel box and pour [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#SDS_running_buffer SDS running buffer] into the negative and positive electrode chamber. | ||
*Remove comp without destroying the gel pocket. | *Remove comp without destroying the gel pocket. | ||
| Line 75: | Line 112: | ||
*Raise amperage up to 20 mA for running the separating gel. | *Raise amperage up to 20 mA for running the separating gel. | ||
*When the distance of the lowest molecular weight standard lane to the gel end is down to 0.5 cm stop the electrophoresis by turning off the power supply. | *When the distance of the lowest molecular weight standard lane to the gel end is down to 0.5 cm stop the electrophoresis by turning off the power supply. | ||
| + | |||
==Polyacrylamide gel staining== | ==Polyacrylamide gel staining== | ||
| - | |||
===Colloidal Coomassie Blue staining=== | ===Colloidal Coomassie Blue staining=== | ||
Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang ''et al''., 2002]. | Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang ''et al''., 2002]. | ||
| - | |||
*agitate the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Colloidal_Coomassie_Brilliant_Blue_G-250_staining_solution staining solution] at 37 °C over night to form the colloids | *agitate the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Colloidal_Coomassie_Brilliant_Blue_G-250_staining_solution staining solution] at 37 °C over night to form the colloids | ||
*After finishing the SDS-PAGE remove gel from gel casting form and tranfer it in to a box. | *After finishing the SDS-PAGE remove gel from gel casting form and tranfer it in to a box. | ||
| Line 87: | Line 123: | ||
*Remove the staining solution | *Remove the staining solution | ||
*wash the gel with dH<sub>2</sub>O | *wash the gel with dH<sub>2</sub>O | ||
| - | *Incubate the gel in ddH<sub>2</sub>O (2- | + | *Incubate the gel in ddH<sub>2</sub>O (2-6 h) for bleaching the background. Shake the gel continuously during incubation. If necessary replace the colored water with new one. |
| + | |||
| + | ===Fairbanks Coomassie staining=== | ||
| + | Modiefid steining protocol from [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf Fairbanks ''et al''., (1971)]. | ||
| + | *After finishing the SDS-PAGE remove gel from gel casting form and tranfer it in to a box with 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution A]. | ||
| + | *Heat the gel in a microwave for 2 min at 600 W. | ||
| + | *Shake the gel for 5-10 minutes. | ||
| + | *Remove solution A and wash the gel with dH<sub>2</sub>O. | ||
| + | *Add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution B] and heat the gel in a microwave for 2 min at 600 W. | ||
| + | *Shake the gel for 5-10 minutes. | ||
| + | *Remove solution B and wash the gel with dH<sub>2</sub>O. | ||
| + | *Add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution C] and heat the gel in a microwave for 2 min at 600 W. | ||
| + | *Shake the gel for 5-10 minutes. | ||
| + | *Remove solution C and wash the gel with dH<sub>2</sub>O. | ||
| + | *For bleaching the background add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution D] and heat the gel in a microwave for 2 min at 600 W. | ||
| + | *Shake the gel for 5-10 minutes. | ||
| + | *Remove solution D and wash the gel with dH<sub>2</sub>O. If necessary repeat the last 3 steps with solution D. | ||
===Silver staining=== | ===Silver staining=== | ||
| - | + | *After finishing the SDS-PAGE remove gel from gel casting form and transfer it in to a box. | |
| - | *After finishing the SDS-PAGE remove gel from gel casting form and | + | |
*Add 50 mL of fixation solution and incubate the gel 1 to 24 h. The formaldehyde (37 %) must be added to the solution short time before. | *Add 50 mL of fixation solution and incubate the gel 1 to 24 h. The formaldehyde (37 %) must be added to the solution short time before. | ||
| - | *Remove the fixation solution and wash the gel 3 times (1-20 min) with 50 mL of 50 % (v/v) ethanol. | + | *Remove the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Silver_staining_solutions fixation solution] and wash the gel 3 times (1-20 min) with 50 mL of 50 % (v/v) ethanol. |
| - | *Add 50 mL of thiosulfate solution and incubate the gel exactly for 1 min. If the gel incubates to long the background becomes to dark. | + | *Add 50 mL of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Silver_staining_solutions thiosulfate solution] and incubate the gel exactly for 1 min. If the gel incubates to long the background becomes to dark. |
*Remove thiosulfate solution and wash the gel 3 times (20 s) with dH<sub>2</sub>O. | *Remove thiosulfate solution and wash the gel 3 times (20 s) with dH<sub>2</sub>O. | ||
| - | *Add 50 mL of impregnation solution and icubate the gel for 15 to 20 min. The formaldehyde (37 %) must be added to the solution short time before. | + | *Add 50 mL of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Silver_staining_solutions impregnation solution] and icubate the gel for 15 to 20 min. The formaldehyde (37 %) must be added to the solution short time before. |
*Remove impregnation solution and wash the gel 2 times (20 s) with dH<sub>2</sub>O. | *Remove impregnation solution and wash the gel 2 times (20 s) with dH<sub>2</sub>O. | ||
| - | *Add 50 mL of | + | *Add 50 mL of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Silver_staining_solutions fixation solution] and incubate the gel until protein bands become visible. |
*Remove the developing solution immediately and wash the gel for 10 to 20 s with dH<sub>2</sub>O. | *Remove the developing solution immediately and wash the gel for 10 to 20 s with dH<sub>2</sub>O. | ||
| - | *Add 50 mL of stop solution and incubate the gel 10 to 20 s. | + | *Add 50 mL of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Silver_staining_solutions stop solution] and incubate the gel 10 to 20 s. |
*Remove the stop solution and wash the gel for 1 min with dH<sub>2</sub>O. | *Remove the stop solution and wash the gel for 1 min with dH<sub>2</sub>O. | ||
==Tryptic digest of gel lanes for analysis with MALDI-TOF== | ==Tryptic digest of gel lanes for analysis with MALDI-TOF== | ||
| - | |||
Note: | Note: | ||
*Make sure to work under a fume hood. | *Make sure to work under a fume hood. | ||
*Do not work with protective gloves to prevent contamination of your sample with platicizers. | *Do not work with protective gloves to prevent contamination of your sample with platicizers. | ||
| - | Reaction tubes have to be cleaned with 60% (v/v) CH<sub>3</sub>CN, 0,1% (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing. | + | Reaction tubes have to be cleaned with 60 % (v/v) CH<sub>3</sub>CN, 0,1 % (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing. |
| - | + | ||
*Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices. | *Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices. | ||
| - | *Gel slices should be washed two times. Therefore add 200 µL 30% (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste. | + | *Gel slices should be washed two times. Therefore add 200 µL 30 % (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste. |
*Dry gel slices at least 30 minutes in a Speedvac. | *Dry gel slices at least 30 minutes in a Speedvac. | ||
*Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation. | *Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation. | ||
| - | *Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C | + | *Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night. |
*Dry gel slices at least 30 minutes in a Speedvac. | *Dry gel slices at least 30 minutes in a Speedvac. | ||
| - | *According to the size of the gel slice, add 5 | + | *According to the size of the gel slice, add 5 - 20 µL 50 % (v/v) ACN / 0,1 % (v/v) TFA. |
| - | *Samples can be used for MALDI measurement or stored at -20 °C. | + | *Samples can be used for MALDI measurement or stored at -20 °C. |
Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH<sub>4</sub>HCO<sub>3</sub> | Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH<sub>4</sub>HCO<sub>3</sub> | ||
| - | *Therefore solubilize lyophilized Trypsin | + | *Therefore solubilize lyophilized [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Used_chemicals Trypsin] in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C. |
| - | == | + | ==Preparation and Spotting for analysis of peptides on Bruker AnchorChips== |
| + | *Spot 0,5 - 1 µL sample aliquot | ||
| + | *Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip. | ||
| + | Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area. | ||
| + | *Dry the prepared spots at room temperature | ||
| + | *Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0.1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial. | ||
| + | *Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots. | ||
| + | |||
| + | ==Bisphenol A analysis== | ||
===Extraction with ethylacetate=== | ===Extraction with ethylacetate=== | ||
| - | * mix 100 µL culture supernatant with 100 µL internal standard ([[Team:Bielefeld-Germany/Protocols#Used_chemicals | bisphenol F | + | * mix 100 µL culture supernatant with 100 µL internal standard ([[Team:Bielefeld-Germany/Protocols/Materials#Used_chemicals | bisphenol F, 100 µg L<sup>-1</sup>]]) |
| - | * add 200 µL [[Team:Bielefeld-Germany/Protocols#Used_chemicals | ethylacetate (HPLC grade)]] for extraction | + | * add 200 µL [[Team:Bielefeld-Germany/Protocols/Materials#Used_chemicals | ethylacetate (HPLC grade)]] for extraction |
* vortex (30 s) | * vortex (30 s) | ||
* centrifuge for phase separation (5 min, 5000 g) | * centrifuge for phase separation (5 min, 5000 g) | ||
| - | * take a bit from upper phase and put it in a clean | + | * take a bit from upper phase and put it in a clean reaction tube |
* SpeedVac at 40 °C to remove ethlyacetate | * SpeedVac at 40 °C to remove ethlyacetate | ||
* solve remaining BPA in water (HPLC grade), vortex (30 s) | * solve remaining BPA in water (HPLC grade), vortex (30 s) | ||
| Line 140: | Line 197: | ||
===HPLC method=== | ===HPLC method=== | ||
* C18 reverse phase column | * C18 reverse phase column | ||
| - | * Isocratic method: 45 % [[Team:Bielefeld-Germany/Protocols#Used_chemicals | Acetonitrile]] | + | * Isocratic method: 45 % [[Team:Bielefeld-Germany/Protocols/Materaials#Used_chemicals | Acetonitrile]] |
* Flow = 0.6 mL min<sup>-1</sup> | * Flow = 0.6 mL min<sup>-1</sup> | ||
* UV-detection at 227 nm | * UV-detection at 227 nm | ||
| - | * Internal standard: 100 mg L<sup>-1</sup> [[Team:Bielefeld-Germany/Protocols#Used_chemicals | Bisphenol F]] (BPF) | + | * Internal standard: 100 mg L<sup>-1</sup> [[Team:Bielefeld-Germany/Protocols/Materials#Used_chemicals | Bisphenol F]] (BPF) |
* Column: | * Column: | ||
** Eurospher II 100-5 C18p by [http://www.knauer.net/ Knauer] | ** Eurospher II 100-5 C18p by [http://www.knauer.net/ Knauer] | ||
| Line 161: | Line 218: | ||
[[File:Bielefeld2011 BPA + Abbauprodukt-2.JPG|900px|left|thumb|Chromatogram obtained using the above described materials and methods. Three distinctive peaks are of interest: BPA at 7,3 min retention time, BPF at 5,8 min retention time and the degradation product of BPA at 4,6 min retention time.]] | [[File:Bielefeld2011 BPA + Abbauprodukt-2.JPG|900px|left|thumb|Chromatogram obtained using the above described materials and methods. Three distinctive peaks are of interest: BPA at 7,3 min retention time, BPF at 5,8 min retention time and the degradation product of BPA at 4,6 min retention time.]] | ||
| - | |||
<br style="clear: both" /> | <br style="clear: both" /> | ||
===LC-ESI-qTOF-MS(-MS)=== | ===LC-ESI-qTOF-MS(-MS)=== | ||
| - | |||
'''HPLC method''' | '''HPLC method''' | ||
* Column: C18 reverse phase column (Knauer [http://beta.knauer.net/products/column-detail-view/productdetail/vertex_plus_column_50_x_2_mm_blueorchid_175_18_c18-1.html Blue Orchid]) | * Column: C18 reverse phase column (Knauer [http://beta.knauer.net/products/column-detail-view/productdetail/vertex_plus_column_50_x_2_mm_blueorchid_175_18_c18-1.html Blue Orchid]) | ||
| Line 201: | Line 256: | ||
* Isolated mass: 243.1 +/- 0.1 | * Isolated mass: 243.1 +/- 0.1 | ||
* Collision energy: 30 eV | * Collision energy: 30 eV | ||
| - | |||
| - | |||
==NAD<sup>+</sup> detection== | ==NAD<sup>+</sup> detection== | ||
| - | |||
| - | |||
===Characterization of molecular beacons=== | ===Characterization of molecular beacons=== | ||
| + | Molecular beacons are nucleic acid probes that fluoresce upon hybridization to a complementary target. To check whether they are suitable for a NAD<sup>+</sup> bioassay involving NAD<sup>+</sup>-dependent DNA ligase from ''Escherichia coli'' (LigA) they can be characterized by thermal profile analysis and determination of the signal-to-background ratio. Detailed background information, tools for the correct design of molecular beacons, a list of synthesis companies as well as protocols for the synthesis and characterization of molecular beacons can be found [http://www.molecular-beacons.com/default.htm here]. The used nucleic acids were designed as follows: | ||
| + | <center> | ||
| + | {|border=1{{Table}} | ||
| + | !Name | ||
| + | !Sequence | ||
| + | !Label | ||
| + | |- | ||
| + | |Molecular beacon | ||
| + | |5'-CCT CTC CGT GTC TTG TAC TTC CCG TCA GAG AGG-3' | ||
| + | |5'-FAM, 3'-Dabcyl | ||
| + | |- | ||
| + | |Target | ||
| + | |5'-GAC GGG AAG TAC AAG ACA C-3' | ||
| + | | | ||
| + | |- | ||
| + | |rowspan="2"|Split target | ||
| + | |5'-TAC AAG ACA C-3' | ||
| + | |5'-phosphate | ||
| + | |- | ||
| + | |5'-GAC GGG AAG-3' | ||
| + | | | ||
| + | |- | ||
| + | |} | ||
| + | </center> | ||
| - | Preferentially, use a complementary oligonucleotide (target) as well as two enshortened oligonucleotides (split target) for | + | The molecular beacon was purchased from [http://www.eurogentec.com/eu-home.html Eurogentec] and labeled with 6-FAM (fluorophor) and Dabcyl (appropriate quencher for 6-FAM). Preferentially, use a complementary oligonucleotide (target) as well as two enshortened oligonucleotides (split target) for characterizing the molecular beacon in its closed and open state. In this way you can verify simultaneously the ineffectiveness of the split target to reach the molecular beacons open state. |
| - | + | ||
* Optimal wavelengths: | * Optimal wavelengths: | ||
** Determine the emission spectrum of your molecular beacon by initially using a literature value for the exctintion wavelength of the labeled fluorophore. For this add 10 µL of 1 µM molecular beacon in 200 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer], mix thoroughly and wait 2 min until measuring the fluorescence with a spectrofluorometer at an defined temperature (optimally 37 °C) . | ** Determine the emission spectrum of your molecular beacon by initially using a literature value for the exctintion wavelength of the labeled fluorophore. For this add 10 µL of 1 µM molecular beacon in 200 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer], mix thoroughly and wait 2 min until measuring the fluorescence with a spectrofluorometer at an defined temperature (optimally 37 °C) . | ||
** Add 10 µL of 2 µM either target or split target and shake at least 6 min until measuring the fluorescence again (when equilibrium is reached). | ** Add 10 µL of 2 µM either target or split target and shake at least 6 min until measuring the fluorescence again (when equilibrium is reached). | ||
| - | ** Repeat the procedure to determine the extinction spectrum using the self-calculated optimal emission wavelength for your fluorophore/quencher combination. For further fluorescence measurements use the extinction wavelength for which the difference between the molecular beacons | + | ** Repeat the procedure to determine the extinction spectrum using the self-calculated optimal emission wavelength for your fluorophore/quencher combination. For further fluorescence measurements use the extinction wavelength for which the difference between the molecular beacons closed state and the open state signals is maximal. |
| - | + | ||
* Thermal profile analysis: | * Thermal profile analysis: | ||
** Prepare a 30 µL reaction mix composed of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] and 1 µM molecular beacon. In a separate sample add 2 µM either target or split target. | ** Prepare a 30 µL reaction mix composed of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] and 1 µM molecular beacon. In a separate sample add 2 µM either target or split target. | ||
** Use a spectrofluorometric thermal cylcer to dertemine the fluorescence as a function of temperature. Start at 80 °C and decrease the temperature stepwise in 1 °C intervals holding each temperature for 1 min and measuring the fluorescence at the end of each step. | ** Use a spectrofluorometric thermal cylcer to dertemine the fluorescence as a function of temperature. Start at 80 °C and decrease the temperature stepwise in 1 °C intervals holding each temperature for 1 min and measuring the fluorescence at the end of each step. | ||
| - | |||
* Signal-to-background ratio (S/B): | * Signal-to-background ratio (S/B): | ||
[[Image:Bielefeld-Germany-2011 MB S-B-ratio.png|200px|thumb|right|Signal-to-background ratio]] | [[Image:Bielefeld-Germany-2011 MB S-B-ratio.png|200px|thumb|right|Signal-to-background ratio]] | ||
| - | |||
**The approach is quite similar to the one that determines the optimal wavelengths which should be used to monitor the fluorescence of the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] (F<sub>buffer</sub>) and the added molecular beacon (F<sub>closed</sub>) as well as the target (F<sub>open</sub>) or split target. Monitor the fluorescence at the optimal wavelengths until the equilibrium is reached each time before you add a new component. | **The approach is quite similar to the one that determines the optimal wavelengths which should be used to monitor the fluorescence of the [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] (F<sub>buffer</sub>) and the added molecular beacon (F<sub>closed</sub>) as well as the target (F<sub>open</sub>) or split target. Monitor the fluorescence at the optimal wavelengths until the equilibrium is reached each time before you add a new component. | ||
| - | ** The signal-to-background ratio can be calculated as shown on the right. Finally, compare the signal-to-background ratios after adding the target and split target to check whether the split target has an effect on the melting of the molecular | + | ** The signal-to-background ratio can be calculated as shown on the right. Finally, compare the signal-to-background ratios after adding the target and split target to check whether the split target has an effect on the melting of the molecular beacons hairpin structure. |
| - | + | ||
* Imaging: | * Imaging: | ||
** Prepare a 200 µL reaction mix composed of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] and 500 nM molecular beacon in a PCR tube. In a separate sample add 600 µM either target or split target. Excitate the samples by a UV transilluminator and take images preferably with a camera that can detect different colours. | ** Prepare a 200 µL reaction mix composed of [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NAD.2B_bioassay_buffer NAD<sup>+</sup> bioassay buffer] and 500 nM molecular beacon in a PCR tube. In a separate sample add 600 µM either target or split target. Excitate the samples by a UV transilluminator and take images preferably with a camera that can detect different colours. | ||
| Line 231: | Line 301: | ||
===Purification of DNA ligase=== | ===Purification of DNA ligase=== | ||
The purification of the overexpressed NAD<sup>+</sup>-dependent DNA ligase gene (<partinfo>K525710</partinfo>) in ''E. coli'' was performed under native conditions and Ni-NTA columns were used utilizing the recombinant protein`s C-terminal 6xHis-tag. | The purification of the overexpressed NAD<sup>+</sup>-dependent DNA ligase gene (<partinfo>K525710</partinfo>) in ''E. coli'' was performed under native conditions and Ni-NTA columns were used utilizing the recombinant protein`s C-terminal 6xHis-tag. | ||
| - | |||
*Cultivation | *Cultivation | ||
| - | **Prepare an overnight culture of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] carrying the plasmid with DNA ligase (<partinfo>K525710</partinfo>) in 30 | + | **Prepare an overnight culture of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] carrying the plasmid with DNA ligase (<partinfo>K525710</partinfo>) in 30 mL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB medium] containing 20 µg mL<sup>-1</sup> chloramphenicol at 37 °C. |
| - | **Dilute the overnight culture in 100 ml [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Autoinduction_medium_for_KRX autoinduction medium] (20 µg mL<sup>-1</sup> chloramphenicol added) to an OD<sub>600</sub>=0.1 and harvest the cells after 4 h growth at 37 °C. | + | **Dilute the overnight culture in 100 ml [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#Autoinduction_medium_for_KRX autoinduction medium] (20 µg mL<sup>-1</sup> chloramphenicol added) to an OD<sub>600</sub> = 0.1 and harvest the cells after 4 h growth at 37 °C. |
**Use the centrifuged cells immediately for protein purification or store them at -20 °C. | **Use the centrifuged cells immediately for protein purification or store them at -20 °C. | ||
| - | |||
*Small-scale purification | *Small-scale purification | ||
**Resuspend a pellet derived from 5 mL cell culture volume in 630 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-10_.28lysis_buffer.29 NPI-10]. | **Resuspend a pellet derived from 5 mL cell culture volume in 630 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-10_.28lysis_buffer.29 NPI-10]. | ||
| Line 247: | Line 315: | ||
**Elute the protein with 300 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-500_.28elution_buffer.29 NPI-500] and centrifuge for 2 min at 890 g (4 °C). Repeat this step and collect flow-through each time. | **Elute the protein with 300 µL [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-500_.28elution_buffer.29 NPI-500] and centrifuge for 2 min at 890 g (4 °C). Repeat this step and collect flow-through each time. | ||
** For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] concentrators ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) with a 5000 molecular weight cut-off PES membrane. Repeat the procedure three times by adding [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#DNA_ligase_buffer DNA ligase buffer] (4 °C) for buffer exchange (remove of imidazole). | ** For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] concentrators ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) with a 5000 molecular weight cut-off PES membrane. Repeat the procedure three times by adding [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#DNA_ligase_buffer DNA ligase buffer] (4 °C) for buffer exchange (remove of imidazole). | ||
| - | ** Check each purification step and especially the purity of the protein in the final condition by [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/ | + | ** Check each purification step and especially the purity of the protein in the final condition by [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Sodium_dodecyl_sulfate_polyacrylamide_gel_electrophoresis_.28SDS-PAGE.29 SDS-PAGE analysis]. |
** Determine the protein concentration with a [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Bradford_Protein_Assay Bradford Protein Assay]. | ** Determine the protein concentration with a [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Bradford_Protein_Assay Bradford Protein Assay]. | ||
** For long-time storage keep the protein at -20 °C. | ** For long-time storage keep the protein at -20 °C. | ||
| - | ** If the binding conditions are not effective enough you can reduce the imidazole concentration in [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-10_.28lysis_buffer.29 NPI-10] to 1-5 mM. If the eluate contains unspecifically bound proteins maybe a washing step with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#NPI-10_.28lysis_buffer.29 NPI-10] containing 50 mM or 100 mM imidazole could help to get higher purity. | + | ** If the binding conditions are not effective enough you can reduce the imidazole concentration in [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-10_.28lysis_buffer.29 NPI-10] to 1-5 mM. If the eluate contains unspecifically bound proteins maybe a washing step with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#NPI-10_.28lysis_buffer.29 NPI-10] containing 50 mM or 100 mM imidazole could help to get higher purity. |
| - | + | ||
*Large-scale purification | *Large-scale purification | ||
| - | ** For production of DNA ligase with high yield see chapter [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#His- | + | ** For production of DNA ligase with high yield see chapter [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Downstream-processing#His-tag_affinity_chromatography His-tag affinity chromatography]. |
===Deadenylation=== | ===Deadenylation=== | ||
| - | |||
The majority of purified DNA ligase from [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] is usually in the adenylated form. But only the apoenzyme is useful for the NAD<sup>+</sup> detection so that the AMP moiety has to be removed. | The majority of purified DNA ligase from [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ ''E. coli'' KRX] is usually in the adenylated form. But only the apoenzyme is useful for the NAD<sup>+</sup> detection so that the AMP moiety has to be removed. | ||
| - | |||
* Mix the purified DNA ligase with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Deadenylation_buffer deadenylation buffer] and incubate for 20 min at 37 °C. | * Mix the purified DNA ligase with [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Deadenylation_buffer deadenylation buffer] and incubate for 20 min at 37 °C. | ||
| - | * For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) concentrators with a 5000 molecular weight cut-off PES membrane. Repeat the procedure several times by adding [https://2011.igem.org/Team:Bielefeld-Germany/Protocols#DNA_ligase_buffer DNA ligase buffer] (remove of NMN and formed NAD<sup>+</sup>). | + | * For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) concentrators with a 5000 molecular weight cut-off PES membrane. Repeat the procedure several times by adding [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Materials#DNA_ligase_buffer DNA ligase buffer] (remove of NMN and formed NAD<sup>+</sup>). |
* Check the deadenylation process by [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Sodium_dodecyl_sulfate_polyacrylamide_gel_electrophoresis_.28SDS-PAGE.29 SDS-PAGE analysis] with high resolution. If the deadenylation was successful there should not be a double band indicating both forms of DNA ligase. | * Check the deadenylation process by [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Sodium_dodecyl_sulfate_polyacrylamide_gel_electrophoresis_.28SDS-PAGE.29 SDS-PAGE analysis] with high resolution. If the deadenylation was successful there should not be a double band indicating both forms of DNA ligase. | ||
* Determine the protein concentration with a [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Bradford_Protein_Assay Bradford Protein Assay]. | * Determine the protein concentration with a [https://2011.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#Bradford_Protein_Assay Bradford Protein Assay]. | ||
| Line 271: | Line 336: | ||
* Monitor the fluorescence with a spectrofluorometer at 37 °C and wait until it reaches equilibrium. | * Monitor the fluorescence with a spectrofluorometer at 37 °C and wait until it reaches equilibrium. | ||
* Add 1 µl NAD<sup>+</sup> in different final concentrations (e.g. 0, 5, 10, 20, 30, 50, 80, 100, 200, 300, 500, 1000, 10000 nM) filling the reaction mix up to 100 µL, stirr shortly and monitor the fluorescence at 37 °C again. | * Add 1 µl NAD<sup>+</sup> in different final concentrations (e.g. 0, 5, 10, 20, 30, 50, 80, 100, 200, 300, 500, 1000, 10000 nM) filling the reaction mix up to 100 µL, stirr shortly and monitor the fluorescence at 37 °C again. | ||
| - | * Determine the initial enhancement rate of | + | * Determine the initial enhancement rate of fluorescence intensity for each NAD<sup>+</sup> concentration (in a range of 200 s after NAD<sup>+</sup> addition). |
* For the calibration curve plot the initial velocity versus the NAD<sup>+</sup> concentration. | * For the calibration curve plot the initial velocity versus the NAD<sup>+</sup> concentration. | ||
| - | Imaging | + | ===Imaging=== |
*It is a similar procedure to the one mentioned above except that you prepare 500 nM molecular beacon and 600 nM target or split target dissolved in bioassay buffer in a PCR tube. | *It is a similar procedure to the one mentioned above except that you prepare 500 nM molecular beacon and 600 nM target or split target dissolved in bioassay buffer in a PCR tube. | ||
*After incubation at 37 °C add 1000 ng DNA ligase and possibly NAD<sup>+</sup> in different concentrations filling the reaction mix up to 200 µL. | *After incubation at 37 °C add 1000 ng DNA ligase and possibly NAD<sup>+</sup> in different concentrations filling the reaction mix up to 200 µL. | ||
*Visualize the fluorescence in a laser scanner or UV transilluminator and document the results. | *Visualize the fluorescence in a laser scanner or UV transilluminator and document the results. | ||
| + | |||
| + | ===Selectivity test=== | ||
| + | * Follow the instructions for the NAD<sup>+</sup> bioassay but use different analytes like NADH, NADP<sup>+</sup>, NADPH, [http://www.sigmaaldrich.com/catalog/ProductDetail.do?D7=0&N5=SEARCH_CONCAT_PNO%7CBRAND_KEY&N4=A5251%7CSIGMA&N25=0&QS=ON&F=SPEC 3-APAD], ATP, ADP or AMP in a final concentration of 100 nM. | ||
| + | |||
| + | ===Coupled enzyme reaction=== | ||
| + | * To check whether the NAD<sup>+</sup> bioassay is suitable for the analysis of NADH-dependent enzymatic reactions it was coupled with the NADH-dependent conversion of pyruvate to L-lactic acid by [http://www.sigmaaldrich.com/catalog/ProductDetail.do?lang=de&N4=59747|SIGMA&N5=SEARCH_CONCAT_PNO|BRAND_KEY&F=SPEC lactic acid dehydrogenase (LDH)] from ''E. coli''. | ||
| + | * Prepare a master mix composed of 250 nM molecular beacon and 250 nM split target (notice that the 5'-end at the nick site has to be phosphorylated) in NAD<sup>+</sup> bioassay buffer (50 mM Tris-HCl pH 8.0, 10 mM MgCl<sub>2</sub>, 2.5 mM CaCl<sub>2</sub>, 5 mM DTT, 0.05 % BSA). | ||
| + | * Incubate for 20 min at 37 °C. | ||
| + | * Meanwhile, prepare a separate reaction mix composed of LDH reaction buffer (20 mM Tris-HCl pH 7.5, 50 mM NaCl), 100 µM NADH and 50 ng/µL LDH. | ||
| + | * Incubate for 5 min at 37 °C. | ||
| + | * Add 650 ng purified and deadenylated LigA to the molecular beacon reaction mix (final concentration 6.5 ng/µL). | ||
| + | * Monitor the fluorescence with a spectrofluorometer at 37 °C and wait until it reaches equilibrium. | ||
| + | * Add 1 µL pyruvate in different final concentrations (e.g. 0, 1, 2.5, 5, 7.5, 10 µM) to the LDH reaction mix. | ||
| + | * Incubate for 2 min at 37 °C. | ||
| + | * Add 1 µL LDH reaction mix to the equilibrated NAD<sup>+</sup> bioassay solution filling the reaction mix up to 100 µL, stirr shortly and monitor the fluorescence at 37 °C again. | ||
| + | * Determine the initial enhancement rate of fluorescence intensity for each pyruvate concentration (in a range of 100 s after LDH reaction mix addition). | ||
| + | * For the calibration curve plot the initial velocity versus the pyruvate concentration. | ||
| + | |||
<br style="clear: both" /> | <br style="clear: both" /> | ||
Latest revision as of 02:05, 29 October 2011


Analytics: This is a list of our used analytical methods.
Bradford Protein Assay
For determination of protein concentrations in protein extracts from bacterial cell lysates or after protein purification steps a Bradford Protein Assay can be applied. For each sample three replicates and at least two dilutions should be measured.
- Prepare differentially concentrated BSA solutions in a range of 0-25 µg/mL for a BSA calibration curve.
- Fill a 96-well plate with 160 µL of each sample.
- Add 40 µL of Bradford [http://www.bio-rad.com/prd/en/US/adirect/biorad?cmd=catProductDetail&vertical=LSR&country=US&lang=en&productID=500-0205 Protein Assay Reagent] to each well and mix thoroughly by stirring as well as pipetting up and down.
- Incubate for 10 min at room temperature.
- Use a plate reader for the measurement of absorbance at 595 nm.
- Calculate the protein concentration with the help of the BSA calibration curve.
Fluorescence measurements
Measuring of [http://partsregistry.org/Part:BBa_E1010 mRFP] with Tecan Infinite® M200 platereader
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 584 nm
- Emission: 620 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
Measuring of [http://partsregistry.org/Part:BBa_J18931 mCitrine] with Tecan Infinite® M200 platereader
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 488 nm
- Emission: 529 nm
- Number of reads: 25
- Manual gain: 75
- Integration time: 20 µs
Measuring of [http://partsregistry.org/Part:BBa_J18931 mCitrine] with Shimadzu RF-5301PC spectrofluorophotometer
- Fill 1 mL sample in a Plastibrand Disposable cuvette 1.5 mL semi-micro PS 12.5 x 12.5 x 45 mm by Brand
- Measure the fluorescence with following settings:
- Exitation: 515 nm
- Emission: 529 nm
- 3 nm exitation filter
- 10 nm emission filter
- sensitivity high or low, depending on fluorescence in sample
Measuring of [http://partsregistry.org/Part:BBa_J18930 mCerulean] with Tecan Infinite® M200 platereader
- Take at least 500 µL sample for each measurement (200 µL is needed for one measurement) so you can perform a repeat determination
- Freeze biological samples at -80 °C for storage, keep cell-free at 4 °C in the dark
- To measure the samples thaw at room temperature and fill 200 µL of each sample in one well of a black, flat bottom 96 well microtiter plate (perform at least a repeat determination)
- Measure the fluorescence in a platereader (we used a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite® M200 platereader]) with following settings:
- 20 sec orbital shaking (1 mm amplitude with a frequency of 87.6 rpm)
- Measurement mode: Top
- Excitation: 433 nm
- Emission: 475 nm
- Number of reads: 25
- Manual gain: 100
- Integration time: 20 µs
Luciferase Measurements
For the luciferase detection we used a [http://www.promega.com/tbs/tb281/tb281.pdf Promega Luciferase Assay System], containing a Cell Culture Lysis Reagent, Luciferase Assay Substrate and Luciferase Assay Buffer
For measuring cells:
- Prepare reaction tubes with 10 µL of high salt buffer (1M K2HPO4, 20mM EDTA, pH 7.8)
- Add 90 µL sample, mix and freeze at -80 °C
- For the measurement thaw by placing the tubes in room temperature water
- Add 300 µL of freshly prepared lysis mix (1X Cell Culture Lysis Reagent, 1.25 mg mL-1 lysozyme, 2.5 mg mL-1 BSA, add water for desired volume)
- Mix and incubate the cells for 10 minutes at room temperature
For measuring cell-free samples start at this point:
- Prepare the Luciferase Assay Reagent, by adding 10 mL of Luciferase Assay Buffer to the vial containing the Luciferase Assay Substrate
- Fill each well of a white, flat bottom 96 well microtiter plate with 20 µL of cell lysate
- For the detection of luciferase use a plate reading luminometer with injector for the Luciferase Assay Reagent and following settings (we used a [http://www.promega.com/b/de/Glomax/glomax_multi_plus.aspx Promega GloMax®-Multi+] Detection System with dual injector):
- Injection volume of Luciferase Assay Reagent: 100 µL
- Delay: 20 secs
- Integration: 3 secs
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
This analytical method can be used for separation and identification of proteins according to their electrophoretic mobility. The mobility is a function of length of the molecular weight. Proteins that have identical charge per unit mass due to binding of SDS results in an equal electrophoretic mobility.
Pouring the polyacrylamide gel
- Make a master mix for the stacking and separating gel without adding ammonium persulfate and TEMED.
- Aliquote 6,5 mL for each separating and 2,5 mL for each stacking gel.
- Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into your gel casting form. Leave about 2 centimeters below the bottom of the comb for the stacking gel.
- Layer isopropanol on top of the gel.
- Leave the separating gel at room temperature for >60 minutes to polymerize.
- Remove isopropanol and wait until the surface is dry.
- Add ammonium persulfate and TEMED to each separating gel aliquote and pour the solution quickly into your gel casting form.
- Insert comb without getting bubbles stuck underneath
- Leave the gel at room temperature for >60 minutes to polymerize.
- For storage
- Remove sealing and store the gel wrapped in moistened paper towel at 4°C.
Preparing the sample
- Mix your protein mixture 4:1 with Laemmli-buffer (30 mL protein solution + 10 mL Laemmli-buffer)
- Heat for 5 minutes at 95 °C.
Running the gel
- Remove sealing, put the polymerized gel into gel box and pour SDS running buffer into the negative and positive electrode chamber.
- Remove comp without destroying the gel pocket.
- Pipet the sample into the gel pockets, adjusting the volume according to the amount of protein in your sample. Make sure to include a lane with molecular weight standards (PageRuler Prestained Protein Ladder™ (Fa. Fermentas)) to determinate the molecular weight of your sample.
- Connect the power lead and run the stacking gel with 10 mA until the blue dye front enters the separating gel.
- Raise amperage up to 20 mA for running the separating gel.
- When the distance of the lowest molecular weight standard lane to the gel end is down to 0.5 cm stop the electrophoresis by turning off the power supply.
Polyacrylamide gel staining
Colloidal Coomassie Blue staining
Modified staining protocol from [http://http://newjournal.kcsnet.or.kr/main/j_search/j_abstract_view.htm?code=B021105&qpage=j_search&spage=b_bkcs&dpage=ar Kang et al., 2002].
- agitate the staining solution at 37 °C over night to form the colloids
- After finishing the SDS-PAGE remove gel from gel casting form and tranfer it in to a box.
- Add 100 mL of the stainig solution to your polyacrylamid gel.
- Incubate the gel in the solution at room temperature until the protein bands got an intensive blue color. Shake the gel continuously during incubation.
- Remove the staining solution
- wash the gel with dH2O
- Incubate the gel in ddH2O (2-6 h) for bleaching the background. Shake the gel continuously during incubation. If necessary replace the colored water with new one.
Fairbanks Coomassie staining
Modiefid steining protocol from [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf Fairbanks et al., (1971)].
- After finishing the SDS-PAGE remove gel from gel casting form and tranfer it in to a box with 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution A].
- Heat the gel in a microwave for 2 min at 600 W.
- Shake the gel for 5-10 minutes.
- Remove solution A and wash the gel with dH2O.
- Add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution B] and heat the gel in a microwave for 2 min at 600 W.
- Shake the gel for 5-10 minutes.
- Remove solution B and wash the gel with dH2O.
- Add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution C] and heat the gel in a microwave for 2 min at 600 W.
- Shake the gel for 5-10 minutes.
- Remove solution C and wash the gel with dH2O.
- For bleaching the background add 100 mL of [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1161216/pdf/biochemj00457-0254.pdf solution D] and heat the gel in a microwave for 2 min at 600 W.
- Shake the gel for 5-10 minutes.
- Remove solution D and wash the gel with dH2O. If necessary repeat the last 3 steps with solution D.
Silver staining
- After finishing the SDS-PAGE remove gel from gel casting form and transfer it in to a box.
- Add 50 mL of fixation solution and incubate the gel 1 to 24 h. The formaldehyde (37 %) must be added to the solution short time before.
- Remove the fixation solution and wash the gel 3 times (1-20 min) with 50 mL of 50 % (v/v) ethanol.
- Add 50 mL of thiosulfate solution and incubate the gel exactly for 1 min. If the gel incubates to long the background becomes to dark.
- Remove thiosulfate solution and wash the gel 3 times (20 s) with dH2O.
- Add 50 mL of impregnation solution and icubate the gel for 15 to 20 min. The formaldehyde (37 %) must be added to the solution short time before.
- Remove impregnation solution and wash the gel 2 times (20 s) with dH2O.
- Add 50 mL of fixation solution and incubate the gel until protein bands become visible.
- Remove the developing solution immediately and wash the gel for 10 to 20 s with dH2O.
- Add 50 mL of stop solution and incubate the gel 10 to 20 s.
- Remove the stop solution and wash the gel for 1 min with dH2O.
Tryptic digest of gel lanes for analysis with MALDI-TOF
Note:
- Make sure to work under a fume hood.
- Do not work with protective gloves to prevent contamination of your sample with platicizers.
Reaction tubes have to be cleaned with 60 % (v/v) CH3CN, 0,1 % (v/v) TFA. Afterwards the solution has to be removed completely followed by evaporation of the tubes under a fume hood. Alternatively microtiter plates from Greiner® (REF 650161) can be used without washing.
- Cut out the protein lanes of a Coomassie-stained SDS-PAGE using a clean scalpel. Gel parts are transferred to the washed reaction tubes/microtiter plate. If necessary cut the parts to smaller slices.
- Gel slices should be washed two times. Therefore add 200 µL 30 % (v/v) acetonitrile in 0,1 M ammonium hydrogen carbonate each time and shake lightly for 10 minutes. Remove supernatant and discard to special waste.
- Dry gel slices at least 30 minutes in a Speedvac.
- Rehydrate gel slices in 15 µL Trypsin-solution followed by short centrifugation.
- Gel slices have to be incubated 30 minutes at room temperature, followed by incubation at 37 °C over night.
- Dry gel slices at least 30 minutes in a Speedvac.
- According to the size of the gel slice, add 5 - 20 µL 50 % (v/v) ACN / 0,1 % (v/v) TFA.
- Samples can be used for MALDI measurement or stored at -20 °C.
Trypsin-solution: 1 µL Trypsin + 14 µL 10 mM NH4HCO3
- Therefore solubilize lyophilized Trypsin in 200 µL of provided buffer and incubate for 15 minutes at 30 °C for activation. For further use it can be stored at -20 °C.
Preparation and Spotting for analysis of peptides on Bruker AnchorChips
- Spot 0,5 - 1 µL sample aliquot
- Add 1 µL HCCA matrix solution to the spotted sample aliquots. Pipet up and down approximately five times to obtain a sufficient mixing. Be careful not to contact the AnchorChip.
Note: Most of the sample solvent needs to be gone in order to achieve a sufficiently low water content. When the matrix solution is added to the previously spotted sample aliquot at a too high water content in the mixture, it will result in undesired crystallization of the matrix outside the anchor spot area.
- Dry the prepared spots at room temperature
- Spot external calibrants on the adjacent calibrant spot positions. Use the calibrant stock solution (Bruker’s “Peptide Calibration Standard II”, Part number #222570), add 125 µL of 0.1% TFA (v/v) in 30% ACN to the vial. Vortex and sonicate the vial.
- Mix the calibrant stock solution in a 1:200 ratio with HCCA matrix and deposit 1 µL of the mixture onto the calibrant spots.
Bisphenol A analysis
Extraction with ethylacetate
- mix 100 µL culture supernatant with 100 µL internal standard ( bisphenol F, 100 µg L-1)
- add 200 µL ethylacetate (HPLC grade) for extraction
- vortex (30 s)
- centrifuge for phase separation (5 min, 5000 g)
- take a bit from upper phase and put it in a clean reaction tube
- SpeedVac at 40 °C to remove ethlyacetate
- solve remaining BPA in water (HPLC grade), vortex (30 s)
- solubility of BPA in water only 300 mg L-1
- for LC-MS analysis of BPA, 300 mg BPA L-1 is definitely enough
- if you want to detect or expect higher concentrations of BPA, solve it in an acetonitrile-water-mix
HPLC method
- C18 reverse phase column
- Isocratic method: 45 % Acetonitrile
- Flow = 0.6 mL min-1
- UV-detection at 227 nm
- Internal standard: 100 mg L-1 Bisphenol F (BPF)
- Column:
- Eurospher II 100-5 C18p by [http://www.knauer.net/ Knauer]
- Dimensions: 150 x 4.6 mm with precolumn
- Particle size: 5 µm
- Pore size: 100 Å
- Material: silica gel
- Software:
- Clarity (Version 3.0.5.505) by [http://www.dataapex.com/ Data Apex]
- Autosampler:
- Midas by [http://www.spark.nl/ Spark Holland]
- Tray cooling: 10 °C
- Pump:
- L-6200A Intelligent Pump by [http://www.hitachi.com/ Hitachi]
- UV-Detector:
- Series 1050 by [http://www.hp.com/ Hewlett Packard]
LC-ESI-qTOF-MS(-MS)
HPLC method
- Column: C18 reverse phase column (Knauer [http://beta.knauer.net/products/column-detail-view/productdetail/vertex_plus_column_50_x_2_mm_blueorchid_175_18_c18-1.html Blue Orchid])
- dimension: 50 x 2 mm
- Pore size: 175 Å
- Particle size: 1.8 µm
- Flow: 0.4 mL min-1
- Column temperature: 30 °C
- Gradient:
- 0 - 1.05 min: 45 % acetonitrile
- 2.55 min: 95 % acetonitrile
- 6.00 min: 95 % acetonitrile
- 6.15 min: 45 % acetonitrile
- 12.00 min: 45 % acetonitrile
- VWR Hitachi LaChrom ULTRA HPLC equipment
- Software: HyStar 3.2, HyStarPP, mircrOTOF Control
Ionization method
- Using Bruker Daltonics micrOTOFQ
- ESI in negative mode
- Mass range: 50 - 1500 m/z
- End plate offset: - 500 V, 107 nA
- Capillary: 2500 V, 4 nA
- Nebulizer: 3 bar
- Dry gas: 8 L min-1
- Quadrupole
- Ion energy: 5 eV
- Low mass: 100 m/z
- Collision energy: 10 eV
- Collision RF: 150 Vpp
- Transfer time: 70 µs
- Pre puls storage: 7 µs
MS-MS
- Isolated mass: 243.1 +/- 0.1
- Collision energy: 30 eV
NAD+ detection
Characterization of molecular beacons
Molecular beacons are nucleic acid probes that fluoresce upon hybridization to a complementary target. To check whether they are suitable for a NAD+ bioassay involving NAD+-dependent DNA ligase from Escherichia coli (LigA) they can be characterized by thermal profile analysis and determination of the signal-to-background ratio. Detailed background information, tools for the correct design of molecular beacons, a list of synthesis companies as well as protocols for the synthesis and characterization of molecular beacons can be found [http://www.molecular-beacons.com/default.htm here]. The used nucleic acids were designed as follows:
| Name | Sequence | Label |
|---|---|---|
| Molecular beacon | 5'-CCT CTC CGT GTC TTG TAC TTC CCG TCA GAG AGG-3' | 5'-FAM, 3'-Dabcyl |
| Target | 5'-GAC GGG AAG TAC AAG ACA C-3' | |
| Split target | 5'-TAC AAG ACA C-3' | 5'-phosphate |
| 5'-GAC GGG AAG-3' |
The molecular beacon was purchased from [http://www.eurogentec.com/eu-home.html Eurogentec] and labeled with 6-FAM (fluorophor) and Dabcyl (appropriate quencher for 6-FAM). Preferentially, use a complementary oligonucleotide (target) as well as two enshortened oligonucleotides (split target) for characterizing the molecular beacon in its closed and open state. In this way you can verify simultaneously the ineffectiveness of the split target to reach the molecular beacons open state.
- Optimal wavelengths:
- Determine the emission spectrum of your molecular beacon by initially using a literature value for the exctintion wavelength of the labeled fluorophore. For this add 10 µL of 1 µM molecular beacon in 200 µL NAD+ bioassay buffer, mix thoroughly and wait 2 min until measuring the fluorescence with a spectrofluorometer at an defined temperature (optimally 37 °C) .
- Add 10 µL of 2 µM either target or split target and shake at least 6 min until measuring the fluorescence again (when equilibrium is reached).
- Repeat the procedure to determine the extinction spectrum using the self-calculated optimal emission wavelength for your fluorophore/quencher combination. For further fluorescence measurements use the extinction wavelength for which the difference between the molecular beacons closed state and the open state signals is maximal.
- Thermal profile analysis:
- Prepare a 30 µL reaction mix composed of NAD+ bioassay buffer and 1 µM molecular beacon. In a separate sample add 2 µM either target or split target.
- Use a spectrofluorometric thermal cylcer to dertemine the fluorescence as a function of temperature. Start at 80 °C and decrease the temperature stepwise in 1 °C intervals holding each temperature for 1 min and measuring the fluorescence at the end of each step.
- Signal-to-background ratio (S/B):
- The approach is quite similar to the one that determines the optimal wavelengths which should be used to monitor the fluorescence of the NAD+ bioassay buffer (Fbuffer) and the added molecular beacon (Fclosed) as well as the target (Fopen) or split target. Monitor the fluorescence at the optimal wavelengths until the equilibrium is reached each time before you add a new component.
- The signal-to-background ratio can be calculated as shown on the right. Finally, compare the signal-to-background ratios after adding the target and split target to check whether the split target has an effect on the melting of the molecular beacons hairpin structure.
- Imaging:
- Prepare a 200 µL reaction mix composed of NAD+ bioassay buffer and 500 nM molecular beacon in a PCR tube. In a separate sample add 600 µM either target or split target. Excitate the samples by a UV transilluminator and take images preferably with a camera that can detect different colours.
Purification of DNA ligase
The purification of the overexpressed NAD+-dependent DNA ligase gene (<partinfo>K525710</partinfo>) in E. coli was performed under native conditions and Ni-NTA columns were used utilizing the recombinant protein`s C-terminal 6xHis-tag.
- Cultivation
- Prepare an overnight culture of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX] carrying the plasmid with DNA ligase (<partinfo>K525710</partinfo>) in 30 mL LB medium containing 20 µg mL-1 chloramphenicol at 37 °C.
- Dilute the overnight culture in 100 ml autoinduction medium (20 µg mL-1 chloramphenicol added) to an OD600 = 0.1 and harvest the cells after 4 h growth at 37 °C.
- Use the centrifuged cells immediately for protein purification or store them at -20 °C.
- Small-scale purification
- Resuspend a pellet derived from 5 mL cell culture volume in 630 µL NPI-10.
- Add 70 µl lysozyme (10 mg/mL) as well as 25 Units benzonase nuclease and mix thoroughly.
- Incubate on ice for 30 min.
- Centrifuge the lysate for 30 min at 12000 g (4 °C). Collect the supernatant (cleared lysate).
- Prepare a [http://www.qiagen.com/products/protein/purification/qiaexpressproteinpurificationsystem/ni-ntaspincolumns.aspx?ShowInfo=1 Ni-NTA spin column] ([http://www.qiagen.com/default.aspx QIAGEN]) by equilibration with 600 µl NPI-10 and centrifugation for 2 min at 890 g (4 °C).
- Load up to 600 µl cleared lysate onto the [http://www.qiagen.com/products/protein/purification/qiaexpressproteinpurificationsystem/ni-ntaspincolumns.aspx?ShowInfo=1 Ni-NTA spin column] and centrifuge for 5 min at 270 g (4 °C). Collect the flow-through.
- Wash the [http://www.qiagen.com/products/protein/purification/qiaexpressproteinpurificationsystem/ni-ntaspincolumns.aspx?ShowInfo=1 Ni-NTA spin column] with 600 µL NPI-20 and centrifuge for 2 min at 890 g (4 °C). Repeat this step and collect flow-through each time.
- Elute the protein with 300 µL NPI-500 and centrifuge for 2 min at 890 g (4 °C). Repeat this step and collect flow-through each time.
- For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] concentrators ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) with a 5000 molecular weight cut-off PES membrane. Repeat the procedure three times by adding DNA ligase buffer (4 °C) for buffer exchange (remove of imidazole).
- Check each purification step and especially the purity of the protein in the final condition by SDS-PAGE analysis.
- Determine the protein concentration with a Bradford Protein Assay.
- For long-time storage keep the protein at -20 °C.
- If the binding conditions are not effective enough you can reduce the imidazole concentration in NPI-10 to 1-5 mM. If the eluate contains unspecifically bound proteins maybe a washing step with NPI-10 containing 50 mM or 100 mM imidazole could help to get higher purity.
- Large-scale purification
- For production of DNA ligase with high yield see chapter His-tag affinity chromatography.
Deadenylation
The majority of purified DNA ligase from [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ E. coli KRX] is usually in the adenylated form. But only the apoenzyme is useful for the NAD+ detection so that the AMP moiety has to be removed.
- Mix the purified DNA ligase with deadenylation buffer and incubate for 20 min at 37 °C.
- For centrifugal ultrafiltration use [http://www.sartorius-stedim.com/DE/en/Centrifugal-Ultrafiltration--Vivaspin-%26-Centrisart/Vivaspin-500/VS0111-VIVASPIN-500-5%2C000-MCWO-PES-25-BOX/6htcki6uex7/m8qc5rabaox/n8uj9p1cyz4/article.htm Vivaspin 500] ([http://www.sartorius.de/index.php?id=156&no_cache=1 Sartorius]) concentrators with a 5000 molecular weight cut-off PES membrane. Repeat the procedure several times by adding DNA ligase buffer (remove of NMN and formed NAD+).
- Check the deadenylation process by SDS-PAGE analysis with high resolution. If the deadenylation was successful there should not be a double band indicating both forms of DNA ligase.
- Determine the protein concentration with a Bradford Protein Assay.
- For long-time storage keep the protein at -20 °C.
NAD+ bioassay
- Prepare a master mix composed of 250 nM molecular beacon and 250 nM split target (notice that the 5'-end at the nick site has to be phosphorylated) in NAD+ bioassay buffer.
- Incubate for 20 min at 37 °C.
- Add 500 ng purified and deadenylated DNA ligase (final concentration 5 ng/µL).
- Monitor the fluorescence with a spectrofluorometer at 37 °C and wait until it reaches equilibrium.
- Add 1 µl NAD+ in different final concentrations (e.g. 0, 5, 10, 20, 30, 50, 80, 100, 200, 300, 500, 1000, 10000 nM) filling the reaction mix up to 100 µL, stirr shortly and monitor the fluorescence at 37 °C again.
- Determine the initial enhancement rate of fluorescence intensity for each NAD+ concentration (in a range of 200 s after NAD+ addition).
- For the calibration curve plot the initial velocity versus the NAD+ concentration.
Imaging
- It is a similar procedure to the one mentioned above except that you prepare 500 nM molecular beacon and 600 nM target or split target dissolved in bioassay buffer in a PCR tube.
- After incubation at 37 °C add 1000 ng DNA ligase and possibly NAD+ in different concentrations filling the reaction mix up to 200 µL.
- Visualize the fluorescence in a laser scanner or UV transilluminator and document the results.
Selectivity test
- Follow the instructions for the NAD+ bioassay but use different analytes like NADH, NADP+, NADPH, [http://www.sigmaaldrich.com/catalog/ProductDetail.do?D7=0&N5=SEARCH_CONCAT_PNO%7CBRAND_KEY&N4=A5251%7CSIGMA&N25=0&QS=ON&F=SPEC 3-APAD], ATP, ADP or AMP in a final concentration of 100 nM.
Coupled enzyme reaction
- To check whether the NAD+ bioassay is suitable for the analysis of NADH-dependent enzymatic reactions it was coupled with the NADH-dependent conversion of pyruvate to L-lactic acid by [http://www.sigmaaldrich.com/catalog/ProductDetail.do?lang=de&N4=59747|SIGMA&N5=SEARCH_CONCAT_PNO|BRAND_KEY&F=SPEC lactic acid dehydrogenase (LDH)] from E. coli.
- Prepare a master mix composed of 250 nM molecular beacon and 250 nM split target (notice that the 5'-end at the nick site has to be phosphorylated) in NAD+ bioassay buffer (50 mM Tris-HCl pH 8.0, 10 mM MgCl2, 2.5 mM CaCl2, 5 mM DTT, 0.05 % BSA).
- Incubate for 20 min at 37 °C.
- Meanwhile, prepare a separate reaction mix composed of LDH reaction buffer (20 mM Tris-HCl pH 7.5, 50 mM NaCl), 100 µM NADH and 50 ng/µL LDH.
- Incubate for 5 min at 37 °C.
- Add 650 ng purified and deadenylated LigA to the molecular beacon reaction mix (final concentration 6.5 ng/µL).
- Monitor the fluorescence with a spectrofluorometer at 37 °C and wait until it reaches equilibrium.
- Add 1 µL pyruvate in different final concentrations (e.g. 0, 1, 2.5, 5, 7.5, 10 µM) to the LDH reaction mix.
- Incubate for 2 min at 37 °C.
- Add 1 µL LDH reaction mix to the equilibrated NAD+ bioassay solution filling the reaction mix up to 100 µL, stirr shortly and monitor the fluorescence at 37 °C again.
- Determine the initial enhancement rate of fluorescence intensity for each pyruvate concentration (in a range of 100 s after LDH reaction mix addition).
- For the calibration curve plot the initial velocity versus the pyruvate concentration.
 "
"