Team:SJTU-BioX-Shanghai/Project/Subproject1/Design
From 2011.igem.org
(Difference between revisions)
(→Design) |
(→Location of Rare Codons) |
||
| (47 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
__NOTOC__ | __NOTOC__ | ||
| - | + | ==Rare tRNA Amount== | |
| - | ==Design== | + | ===Design=== |
| - | + | ||
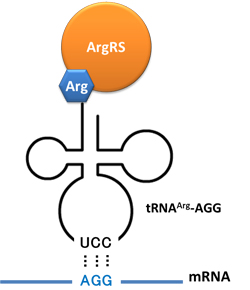
| - | + | In this part we have overexpressed rare tRNA<sup>Arg</sup>-AGG in the cell. The rare tRNA can recognize AGG codon on the mRNA. | |
| - | ''' | + | '''tRNA<sup>Arg</sup>-AGG''': tRNA<sup>Arg</sup>-AGG is over expressed under the control of trc promoter (induced by IPTG). |
| - | ''' | + | This rare tRNA<sup>Arg</sup> can be charged with Arg by native '''Arginyl-tRNA Synthetase(ArgRS)''' in E.coli. |
| + | [[image:11SJTU_Rare_03.jpg|center|tRNA Modulator]] | ||
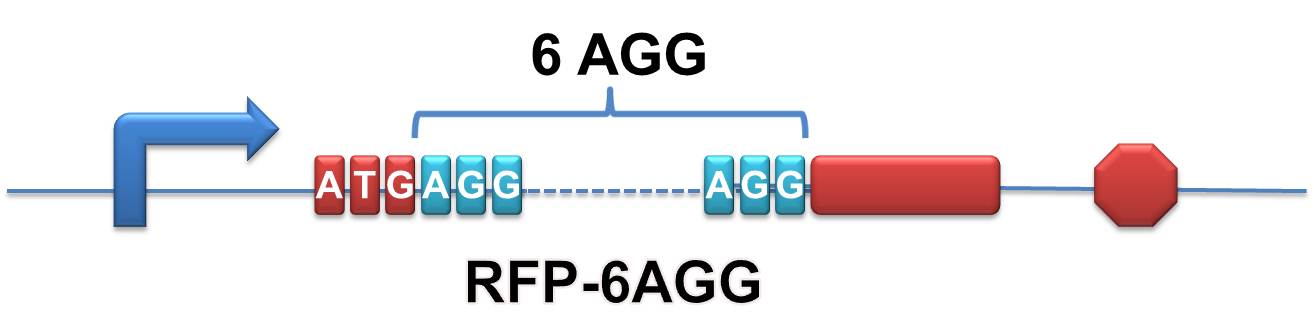
| + | '''RFP-6AGG''': we have inserted 6 AGG codons after the start codon ATG in the RFP gene. | ||
| - | + | [[image:11SJTU_rare_00.jpg|500px|center]] | |
| - | === | + | ===Action=== |
| - | + | When rare tRNA<sup>Arg</sup>-AGG is not over-expressed, RFP expression is hindered. When tRNA<sup>Arg</sup>-AGG is over-expressed, this tRNA can recognize the AGG codon on the mRNA so a large amount of RFP is produced. | |
| - | + | [[image:11SJTU_rare_01.jpg|500px|center]] | |
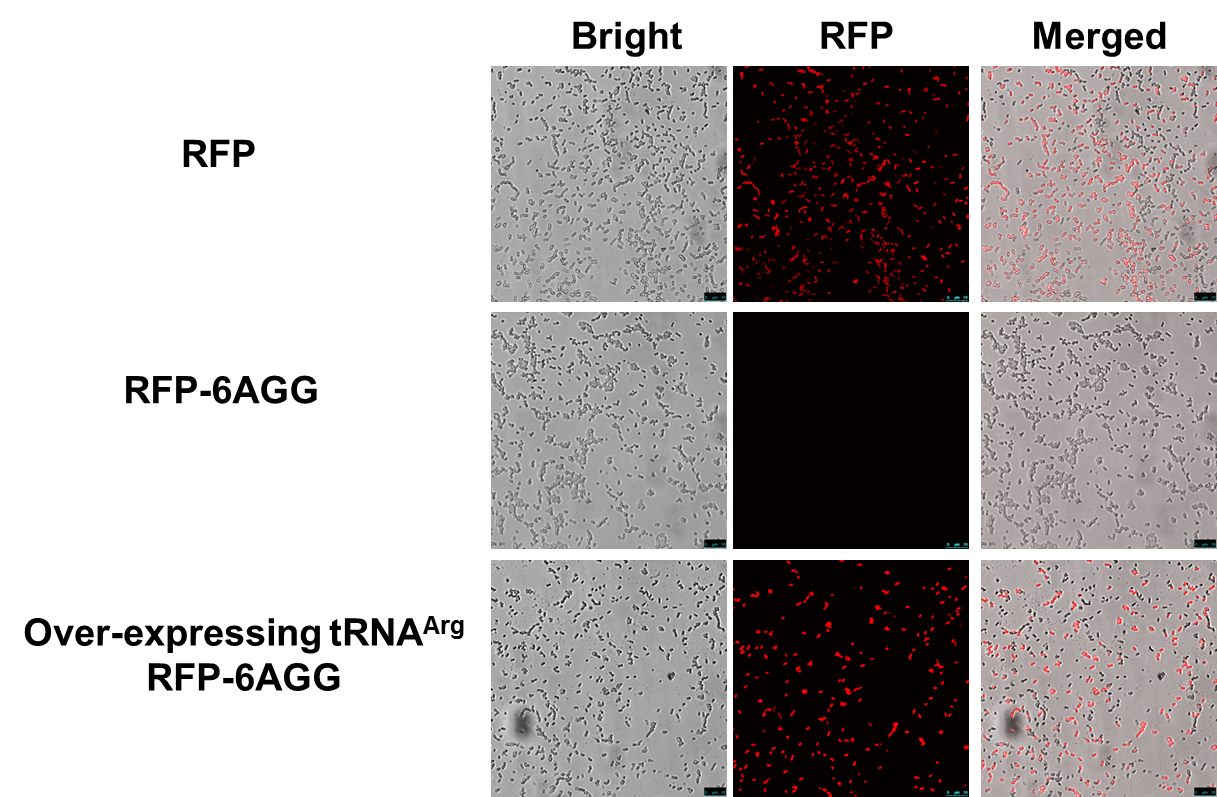
| - | + | ===Result=== | |
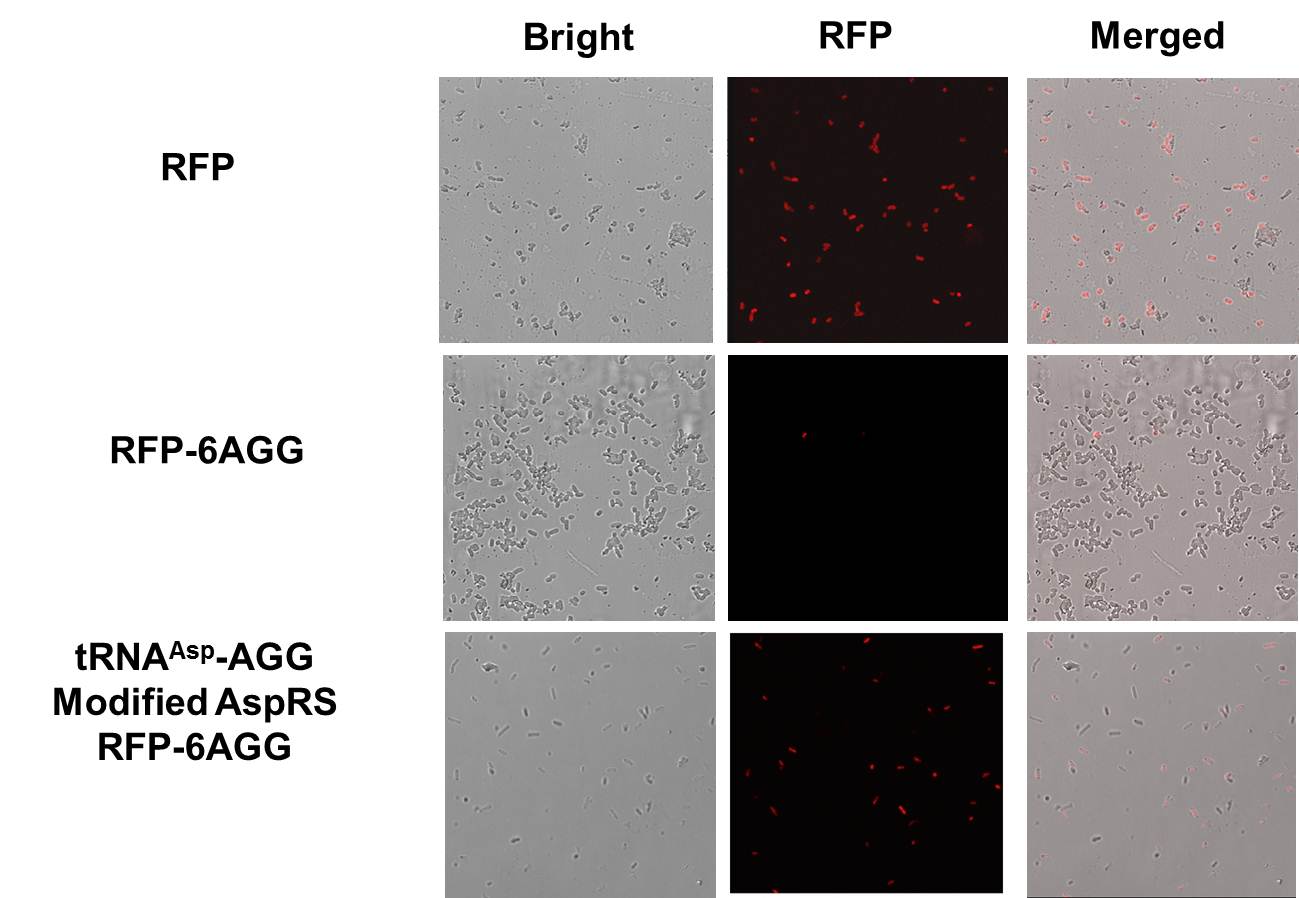
| - | + | [[File:11SJTU_rare_02.png|thumb|500px|center|''Fig 1''. Confocal Microscope examining RFP expression. '''RFP has been largely produced in cells overexpressing tRNAArg-AGG.''']] | |
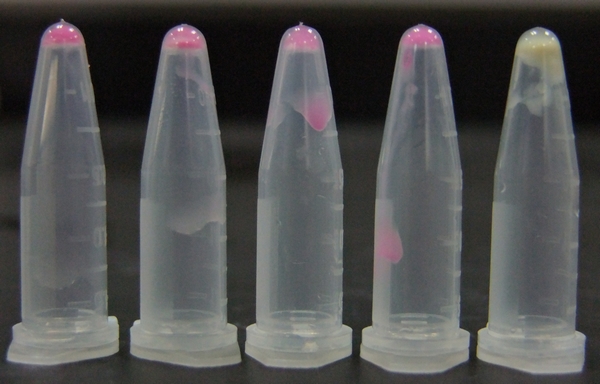
| - | ''' | + | [[File:11SJTU_rare_10.jpg|thumb|500px|center|''Fig 2'' Cells over-expressing tRNAArg-AGG emit bright red fluorescence as wild type RFP (first one from the left). Control (first one from the right) exhibits no red fluorescence.]] |
| - | + | RFP has been largely produced in cells overexpressing tRNA<sup>Arg</sup>. No RFP can be observed in cells without rare tRNA overexpression. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | RFP has been largely produced in cells overexpressing | + | |
'''We have successfully controlled protein expression by controlling rare tRNA amount.''' | '''We have successfully controlled protein expression by controlling rare tRNA amount.''' | ||
| - | + | ==aaRS== | |
| - | + | ===Design=== | |
We control the translation process by modifying the tRNA and aaRS that are originally not for Arg. | We control the translation process by modifying the tRNA and aaRS that are originally not for Arg. | ||
| - | + | tRNA<sup>Asp</sup>-AGG: tRNA<sup>Asp</sup> with its anticodon mutated to CCU can base pair with rare codon AGG, which is originally the codon for Arg. This tRNA is under the constitutive aspV promoter. | |
TDRS: Aspartyl aminoacyl tRNA synthetase (AspRS) without anticodon recognition domain under the control of T7 promoter and lac operator. To deprive AspRS of its anticodon specificity, we analyzed the structure of AspRS and expressed a truncated AspRS without anticodon recognition domain. This modified enzyme keeps its ability of aminoacylation while loses its activity of recognizing anticodon of tRNA. | TDRS: Aspartyl aminoacyl tRNA synthetase (AspRS) without anticodon recognition domain under the control of T7 promoter and lac operator. To deprive AspRS of its anticodon specificity, we analyzed the structure of AspRS and expressed a truncated AspRS without anticodon recognition domain. This modified enzyme keeps its ability of aminoacylation while loses its activity of recognizing anticodon of tRNA. | ||
| Line 68: | Line 57: | ||
Luciferase-4AGG: luciferase with 4AGG insertions | Luciferase-4AGG: luciferase with 4AGG insertions | ||
| - | + | ===Action=== | |
| - | When the modified enzyme is produced under induction, it can charge | + | When the modified enzyme is produced under induction, it can charge tRNA<sup>Asp</sup>-AGG with Arg. Then the charged tRNA can get through the rare codons on the mRNA, so that RFP or luciferase can be produced. |
| - | + | ===Result=== | |
| - | + | [[image:11SJTU_rare_05.jpg|thumb|500px|center|''Fig 2''. With our device (BBa_K567012 and BBa_K567011), RFP-6AGG is expressed.]] | |
| - | + | [[image:11SJTU-ZBresult2.JPG|600px|frame|center|''Fig 3.'' aaRS Modulator + Reporter for Qualitative Analysis. aaRS Modulator + RFP-6AGG''(the middle three)'' emit red fluorescence. Wild type RFP ''(the first one from the left)'' exhibits bright red fluorescenceis. Control ''(first one from the right)'' exhibits no red fluoresence.]] | |
| - | + | We have used PT7-RFP-6AGG (BBa_K567017) as our Reporter. We have constructed tRNA<sup>Asp</sup>-AGG and PT7-TDRS (AspRS without anticodon recognition domain) (BBa_K567012 and BBa_K567011). tRNA<sup>Asp</sup>-AGG, which can recognize rare codon AGG, is under constitutive promoter. '''With our device, RFP is successfully produced. Without our device, little RFP is observed. ''' | |
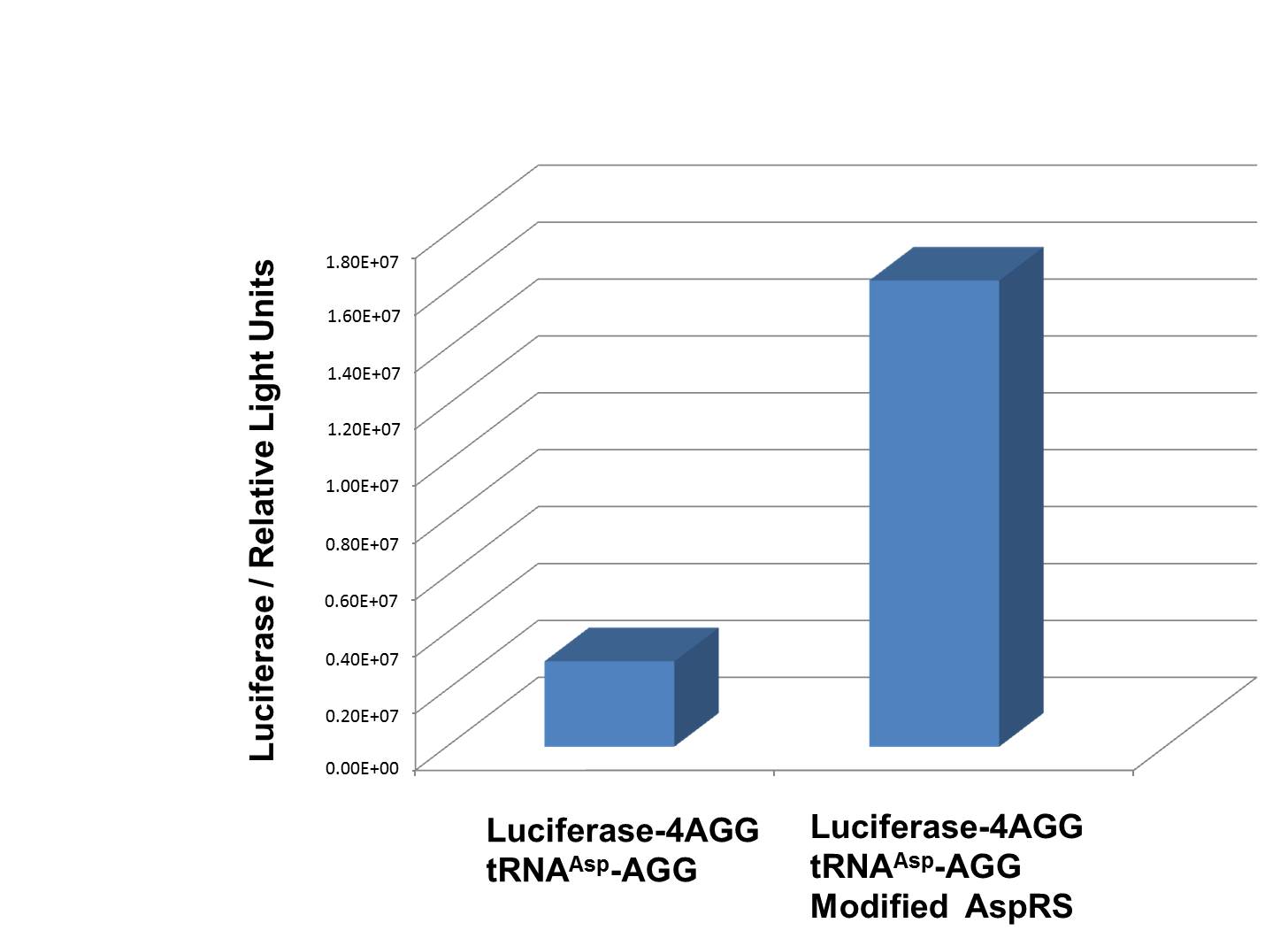
| - | We have used PT7-Luc-4AGG (BBa_K567009) as our Reporter to test the function of PT7-TDRS (BBa_K567011) and | + | [[image:11SJTU_rare_04.jpg|thumb|500px|center|''Fig 1''. Examination of luciferase production with and without device. ER2566 cannot produce luciferase with PT7-Luc-4AGG (BBa_K567009) only. When tRNAAsp-AGG (BBa_K567012) and PT7-TDRS (BBa_K567011) are co-transformed into the cell, luciferase production is increased. The results proved that aaRS can regulate protein biosynthesis. ]] |
| + | |||
| + | We have used PT7-Luc-4AGG (BBa_K567009) as our Reporter to test the function of PT7-TDRS (BBa_K567011) and tRNA<sup>Asp</sup>-AGG (BBa_K567012). Results are shown above. '''Luciferase production has been largely increased with our device.''' | ||
| + | |||
| + | The function of Switch is characterized by the amount of luciferase expressed. The amount of luciferase expressed is reflected by the light emitted when luciferase acts on the appropriate luciferin substrate. The light can be measured by luminometer and the quantity is positively correlated with the amount of luciferase and its activity (learn more...). | ||
'''We successfully controlled protein expression by manipulating aaRS.''' | '''We successfully controlled protein expression by manipulating aaRS.''' | ||
| - | + | ==Number of Rare Codons== | |
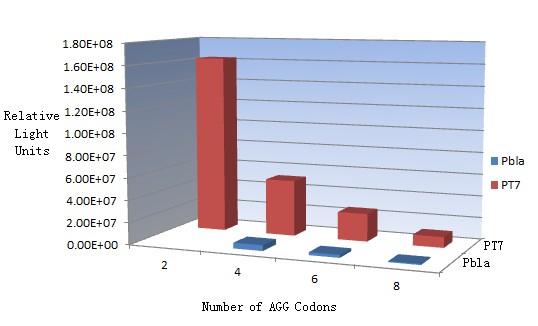
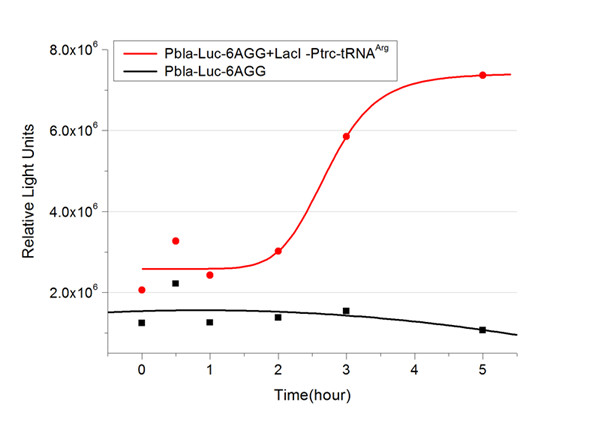
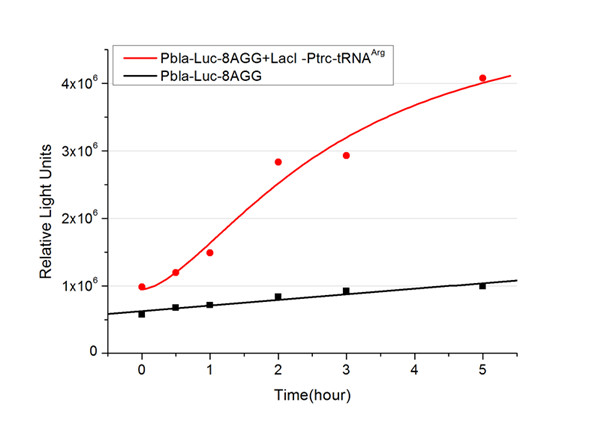
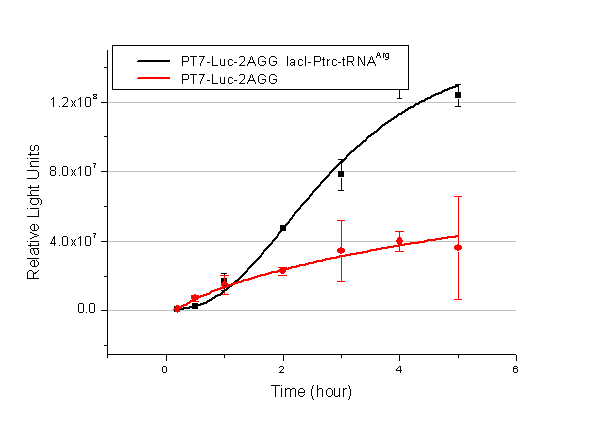
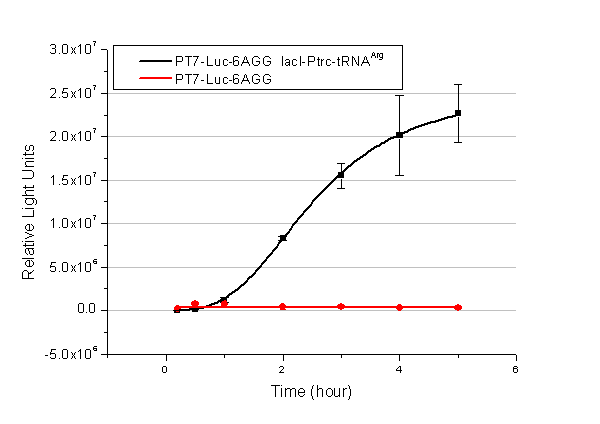
In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or bla promoter[1] are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation[1]. | In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or bla promoter[1] are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation[1]. | ||
| Line 95: | Line 88: | ||
A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase | A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase | ||
| - | + | ===Influence of inserted AGG codon number=== | |
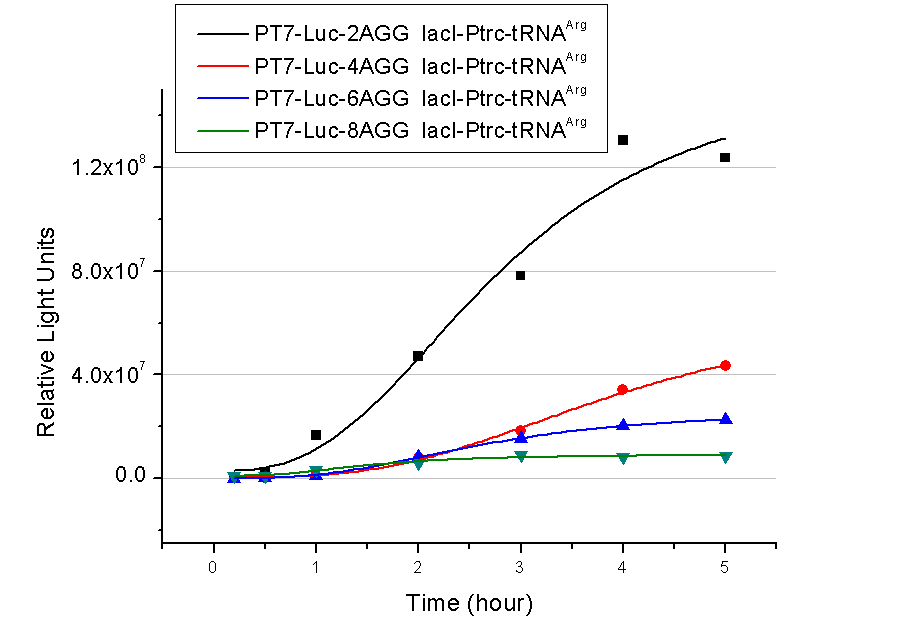
| - | + | The influence of different number of rare codons in regulating protein biosynthesis is shown below: | |
| - | + | [[image:11SJTU_rare_t7.png|500px|center]] | |
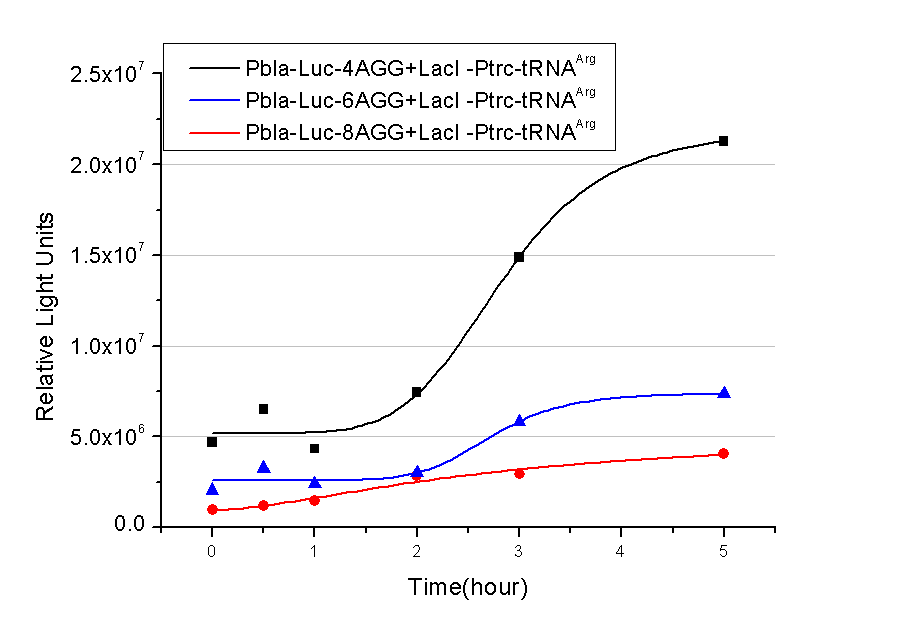
| + | [[image:11SJTU_rare_bla.png|500px|center]] | ||
| - | Results show that the more rare codons are inserted, the lower the background expression and the narrower the range our device can regulate | + | Results show that the more rare codons are inserted, the lower the background expression and the narrower the range our device can regulate. |
| - | + | [[image:11SJTUzhuzhuangtu-compare.jpg|frame|center|''Fig.7'' Enzyme activity of luciferase reaching plateau phase.]] | |
This picture reflects more clearly that the more rare codons are inserted, the lower the background expression and the narrower the range of device regulation. We are able to predict the outcome of influence of different number of rare codons in protein biosynthesis, offering valuable information for device usage. | This picture reflects more clearly that the more rare codons are inserted, the lower the background expression and the narrower the range of device regulation. We are able to predict the outcome of influence of different number of rare codons in protein biosynthesis, offering valuable information for device usage. | ||
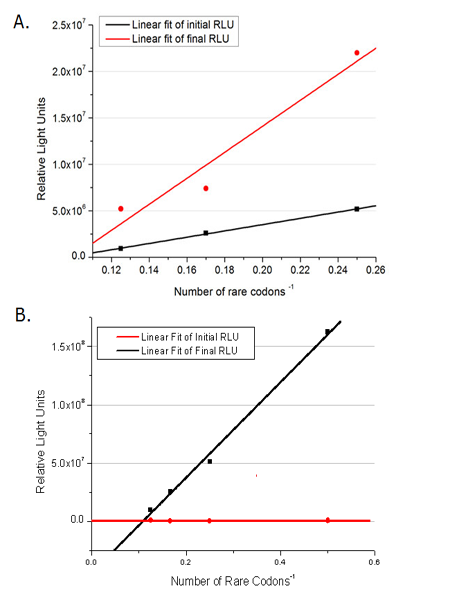
| - | + | [[image:11sjtu_Compare_DATA_Fitting.png|frame|center|''Fig.11'' The comparison between background expression and induced expression of luciferase with different rare codon insertions. (A)PT7-luc reporters. (B) Pbla-luc reporters]] | |
| + | |||
| + | ===Influence of different strengths of target protein promoters=== | ||
We examined the influence of different Reporter promoters on the working curve of our device, which is reflected by luciferase activity. The working range of our device is pre-defined by the strength of target protein promoter, T7 promoter and bla promoter in our project. | We examined the influence of different Reporter promoters on the working curve of our device, which is reflected by luciferase activity. The working range of our device is pre-defined by the strength of target protein promoter, T7 promoter and bla promoter in our project. | ||
| - | + | [[image:11sjtu_Compare_8.png|frame|center|''Fig.8'' (A) Working curve of tRNA Modulator ''lacI''-Ptrc-tRNA<sup>Arg</sup> under Reporter P''bla''-Luc-8AGG reflected by bioluminescence emitted from the luciferin reaction. (B) Working curve of tRNA Modulator under Reporter PT7-Luc-8AGG. Here we analyze the influences of strong/weak promoter in the working curve of tRNA Modulator. | |
| + | Moreover, strong promoter (T7) of target gene can improve the titration curve of tRNA Modulator, indicating that tRNA Modulator works better under strong target protein promoters. ]] | ||
| + | |||
| + | ===Note: Our device can be used as a regulating tool=== | ||
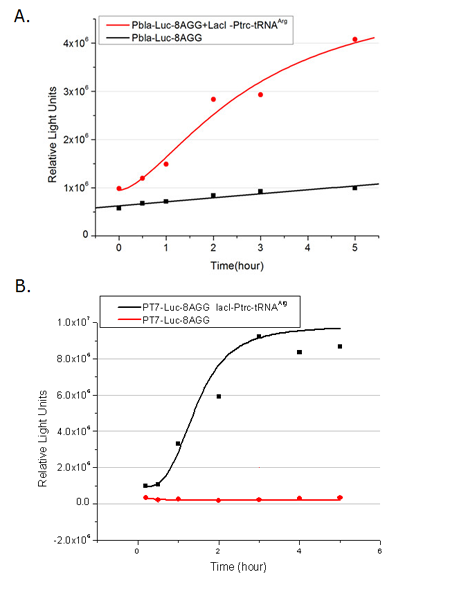
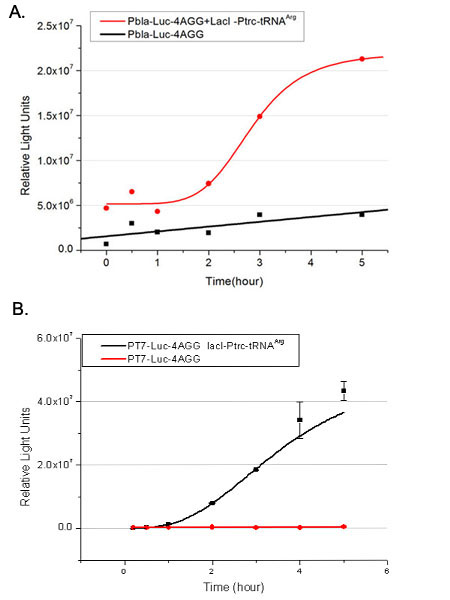
We have tested luciferin reaction in cells. We examined the changes in luciferase enzyme activity over time after rare tRNA expression is induced. The amount of luciferase is reflected indirectly by the bioluminescence emitted from the luciferin reaction. Results are shown below: | We have tested luciferin reaction in cells. We examined the changes in luciferase enzyme activity over time after rare tRNA expression is induced. The amount of luciferase is reflected indirectly by the bioluminescence emitted from the luciferin reaction. Results are shown below: | ||
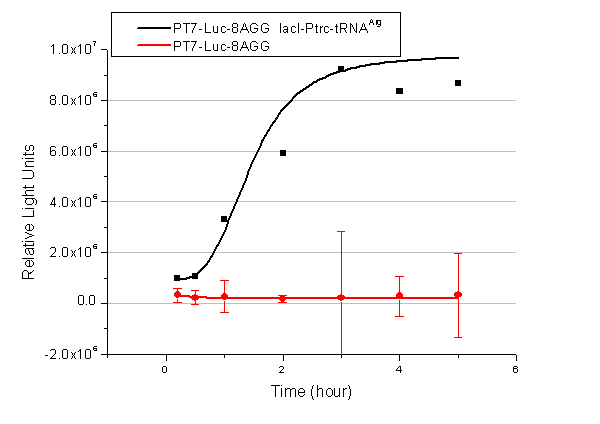
| - | + | [[image:11-SJTU-compare-4.jpg|frame|center|''Fig.1'' (A)Enzyme activity of luciferase shown by bioluminescence emitted from the luciferin reaction reflecting the working curve of tRNA Modulator ''lacI''-Ptrc-tRNA<sup>Arg</sup> ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567001 BBa_K567001]) under Reporter P''bla''-Luc-4AGG([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567006 BBa_K567006]). (B)Enzyme activity of luciferase shown by bioluminescence emitted from the luciferin reaction reflecting the working curve of tRNA Modulator lacI-Ptrc-tRNA<sup>Arg</sup> ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567001 BBa_K567001]) under Reporter PT7-Luc-4AGG ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567009 BBa_K567009]). The working curve of tRNA Modulator ''lacI''-Ptrc-tRNA<sup>Arg</sup> fits typical titration curve.]] | |
Here we use the above two curves as examples to characterize the working curve of our device. Both curves fits typical titration curve, indicating that our device can function as a regulating tool. | Here we use the above two curves as examples to characterize the working curve of our device. Both curves fits typical titration curve, indicating that our device can function as a regulating tool. | ||
| Line 121: | Line 120: | ||
The rest of the working curves are shown here: | The rest of the working curves are shown here: | ||
| - | + | <gallery caption="Pbla"> | |
| + | image:11SJTUResultPlbla6.jpg|''Fig.2'' | ||
| + | image:11SJTUResultPbla8.jpg|''Fig.3'' | ||
| + | </gallery> | ||
| + | |||
| + | <gallery caption="PT7"> | ||
| + | image:11SJTUT7-2.png|''Fig.4'' | ||
| + | image:11SJTUT7-6.png|''Fig.5'' | ||
| + | image:11SJTUT7-8.png|''Fig.6'' | ||
| + | </gallery> | ||
| + | '''Note''':Click to see large figures. | ||
Results showed that all the devices’ working curves fit titration curve, indicating that our device can act as a satisfying regulating tool. | Results showed that all the devices’ working curves fit titration curve, indicating that our device can act as a satisfying regulating tool. | ||
| - | + | From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced lacI-Ptrc-tRNA<sup>Arg</sup> (BBa_K567001) compared with UV excitation induced sulA promoter-tRNA<sup>Arg</sup>(BBa_K567002), though sulA promoter-tRNA<sup>Arg</sup> responded quicker to signals. So in the above experiments, we test with lacI-Ptrc-tRNA<sup>Arg</sup>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced lacI-Ptrc- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | ==Location of Rare Codons== |
| - | + | ||
Latest revision as of 05:33, 28 October 2011
 "
"