Team:British Columbia/Accomplishments
From 2011.igem.org
| Line 16: | Line 16: | ||
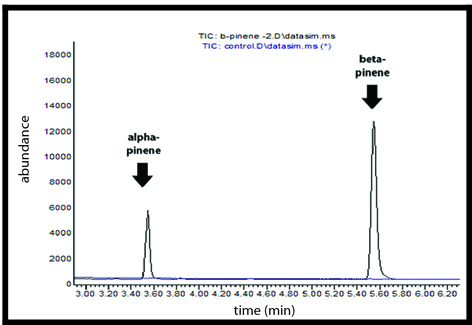
<b></b><font size="1"><h3>GC-MS Chromatogram</h3></font> | <b></b><font size="1"><h3>GC-MS Chromatogram</h3></font> | ||
| - | + | [[File:UBCiGEM_GC_beta_pinene.jpg]] | |
Revision as of 03:20, 28 October 2011

Wet Laboratory Achievements
1. In vitro assay production of monoterpene in bacteria
Geranyl Pyrophosphate (GPP) Assay
Alpha-pinene and Beta-pinene synthases were purified using His SpinTrap Ni-affinity columns and were assayed in vitro with GPP. Using gas chromatography-mass spectrometry (GCMS), we confirmed the synthesis of alpha and beta pinene monoterpenes from our enzyme assays.
GC-MS Chromatogram
2. Diterpene production in yeast
3. Mountain Pine Beetle & Yeast Co-culture
To prove that release methods by incubation of Mountain Pine Beetles
3. Blue Stain Fungi & Yeast Co-culture Co-culture
Modeling Achievements
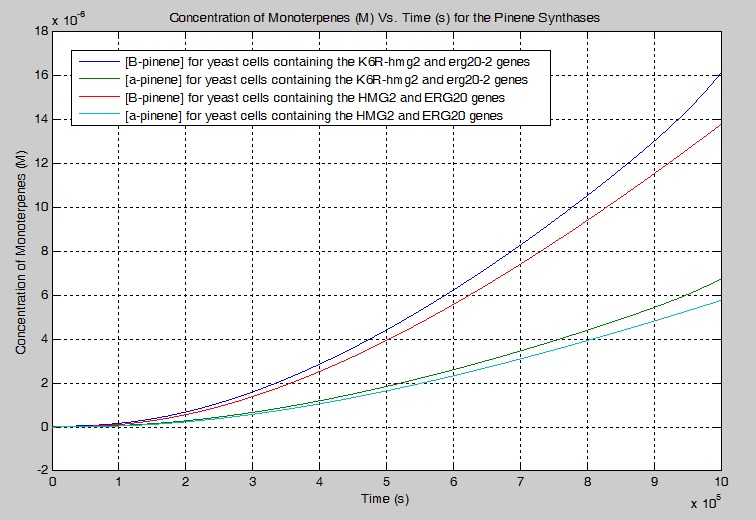
Monoterpene Production
We modeled a simplified and modified version of the mevalonate pathway that describes our engineered yeast cells. We created a series of differential equations to model each chemical reaction that were based on first-order Michaelis-Menten kinetics. Enzyme constants were estimated/found using literature values and the simulations were conducted using MATLAB.
Our simulation showed that 17.10% more beta-pinene is produced and 17.00 % more alpha-pinene is produced when K6R-hmg2 and erg20-2 are used instead of HMG2 and ERG20 alone in a yeast cell. For manufacturing purposes, sensitivity analysis was performed and it was determined that the pathway could be improved to increase the production of monoterpenes by increasing the concentration of the enzymes K6R-HMG2 and IDI1 for DMA-PP. In particular, the tripling the [K6R-HMG2] increases the [GPP] by 8.7000 times. The tripling the [K6R-HMG2] and [IDI1] for the production of DMA-PP increases the [B-pinene] by 7.3166 times and 1.3052 times respectively.
We simulated the expansion of the MPB population from year 2011 to 2020 using the estimates obtained from the clustering analysis. For cost estimation and prediction of emergence of subpopulations, refer to our Model Methodology above.
 "
"