Team:British Columbia/Notebook/Week 16
From 2011.igem.org
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Notebook}} | {{Template:Notebook}} | ||
| - | + | <html> | |
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b>Week 16: Sep 18-28</b><html><a name="w16"></a></html> | ||
| - | ==The week before the wiki-freeze== | + | ===The week before the wiki-freeze=== |
All is busy in the lab. Daisy, Jacob and Joe are working on characterizing the monoterpene synthases in C41 bacteria. Joe is working on characterizing the GAL1 promoter from the registry. Gurpal is trying to get data for his ERG20 and erg20-2 parts. Check out our <html><a href="https://2011.igem.org/Team:British_Columbia/Data">data page</a></html> for results! | All is busy in the lab. Daisy, Jacob and Joe are working on characterizing the monoterpene synthases in C41 bacteria. Joe is working on characterizing the GAL1 promoter from the registry. Gurpal is trying to get data for his ERG20 and erg20-2 parts. Check out our <html><a href="https://2011.igem.org/Team:British_Columbia/Data">data page</a></html> for results! | ||
| Line 68: | Line 71: | ||
[[File:ubcigemhoodies.jpg | center ]] | [[File:ubcigemhoodies.jpg | center ]] | ||
| - | |||
| - | [[ | + | ===Poster and Presentation=== |
| + | |||
| + | The team is busy working on their poster and presentation. | ||
| + | |||
| + | |||
| + | ===aGEM=== | ||
| + | Joanne and Jacob went to aGEM, a pre-jamboree iGEM meetup based in Alberta. Jacob had the opportunity to give UBC's presentation to a group of judges and the students from the Alberta teams. He received some great feedback about the project and presentation, saw the fantastic presentations from the Alberta teams, and had the opportunity to chat with the judges about the future of synthetic biology. It was an amazing experience! | ||
| + | |||
| + | |||
| + | ===Expression of Beta-pinene bacteria=== | ||
| + | |||
| + | Daisy purified the pellets of beta-pinene that Joe prepare a week ago and store at -80C. She is certain that there is beta-pinene in these cells because previous SDS-page data shows a strong band slightly below 70kDa ( the size of both alpha and beta-pinene synthases). Well, she performed a GPP assay and prepared those samples for GC-MS , and surprising it came out that the beta-pinene synthase was producing alpha-pinene and mostly beta-pinene: | ||
| + | |||
| + | [[Image:Ubcigem2011betapingcms.jpg | center | frame | Sample GC-MS data for our beta-pinene synthase as expressed by C41 ''E. coli'']] | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 04:33, 27 October 2011

 |
 |
 |
 |
 |
Contents |
The week before the wiki-freeze
All is busy in the lab. Daisy, Jacob and Joe are working on characterizing the monoterpene synthases in C41 bacteria. Joe is working on characterizing the GAL1 promoter from the registry. Gurpal is trying to get data for his ERG20 and erg20-2 parts. Check out our data page for results!
Expression of Alpha-pinene, Beta-pinene,1,8 Cineole in bacteria
Daisy, Joe and Jacob decided to characterize the synthases in C41 bacteria. This time, they were going to not evaporate any of the pentane. It was suggested that evaporating the pentane, while reduces the volume, also evaporates away some of the product as they are very volatile. This is particularly true of alpha-pinene and beta-pinene.
There were several methods that could be tried. Daisy talked to Chris Keeling and he says that they do the in vitro assay in the GCMS vials (the 2 mL vials). This way, they do not have to transfer the pentane.
The other method was to try in a smaller glass vial, such as the yeast preparation vials.
Daisy and Joe decided that because we have four samples:
- alpha-pinene - done in yeast preparation vials (can hold larger volume)
- beta-pinene - done in GCMS vials
- 1,8 - cineole - done in yeast preparation vials
- 1,8-cineole - done in GCMS vials
In addition, some of the samples throughout the preparation to the GCMS were taken and run on an SDS-PAGE to confirm protein expression.
Some of the problems with this batch was that the amount of time it took to grow to absorbance 0.8 was much longer than usual. This may be because the usual incubator at 37 degrees was not available. Joe instead had to use the 38-39 (slight variation in temperature) incubator. It look a couple of hours longer to grow to that temperature.
Daisy asked quite a few people to see if they knew whether growing it at a higher temperature would affect enzyme function or inclusion body formation. Most people could not give an definitive answer as to whether it would severely impact enzyme production. Regardless, growing bacteria at 38 to 39 is not great. However, due to the amount of time we have, we decided to proceed with the pellets anyway.
The results came back:
- alpha-pinene - GCMS data
- beta-pinene - no data
- 1,8 cineole - no data
- 1,8 cineole - no data
alpha-pinene and beta-pinene were grown at the same time in the same batch with the same incubator. The only difference was the way the in vitro assay was set up. The alpha-pinene was set up in a yeast preparation vial with a larger volume of enzyme as opposed to beta-pinene which was done at a much smaller volume.
Beta-pinene was later re-done with the yeast vial preparation method for the in vitro assay.
The protein samples were run on an SDS PAGE 10% gel. The gels were ran as:
- Pre-induction alpha-pinene
- Post-induction alpha-pinene
- Alpha-pinene purified protein
- Pre-induction beta-pinene
- Post-induction beta-pinene
- Beta-pinene purifed protein
The gels were run properly and there were some results but somewhat unclear. Two Bohlmann Lab members (thanks for letting Daisy bother you guys!!) helped Daisy troubleshoot the gel and try to interpret the results. It turns out that because the protein was expressed in bacteria and the cell lysate was run though a gravity filtered nickel column (his tag), there are some other bacterial proteins that also bind to the column. When we ran the protein on the gel, we saw several bands in the purified protein lane.
The two Bohlmann lab members also suggested doing a western with anti-his antibodies but we do not have any anti-his antibodies and neither do they.
Derek helped Daisy take a picture of the protein gels and also suggested a western.
We also tried the yeast expression protocols again without the evaporation and there were no results.
Gurpal is slaving away over Model 1. Joe, Jacob and Sam are cracking on Model 2. Shing is advising!
Laura, together with many members of the team and graduate advisors, delivered an inspiring workshop to high school students at the Vancouver Science World, read more here!
We've also just shipped our 6 well documented parts to iGEM HQ!!
And received our awesome team hoodies from CustomInk!!
Poster and Presentation
The team is busy working on their poster and presentation.
aGEM
Joanne and Jacob went to aGEM, a pre-jamboree iGEM meetup based in Alberta. Jacob had the opportunity to give UBC's presentation to a group of judges and the students from the Alberta teams. He received some great feedback about the project and presentation, saw the fantastic presentations from the Alberta teams, and had the opportunity to chat with the judges about the future of synthetic biology. It was an amazing experience!
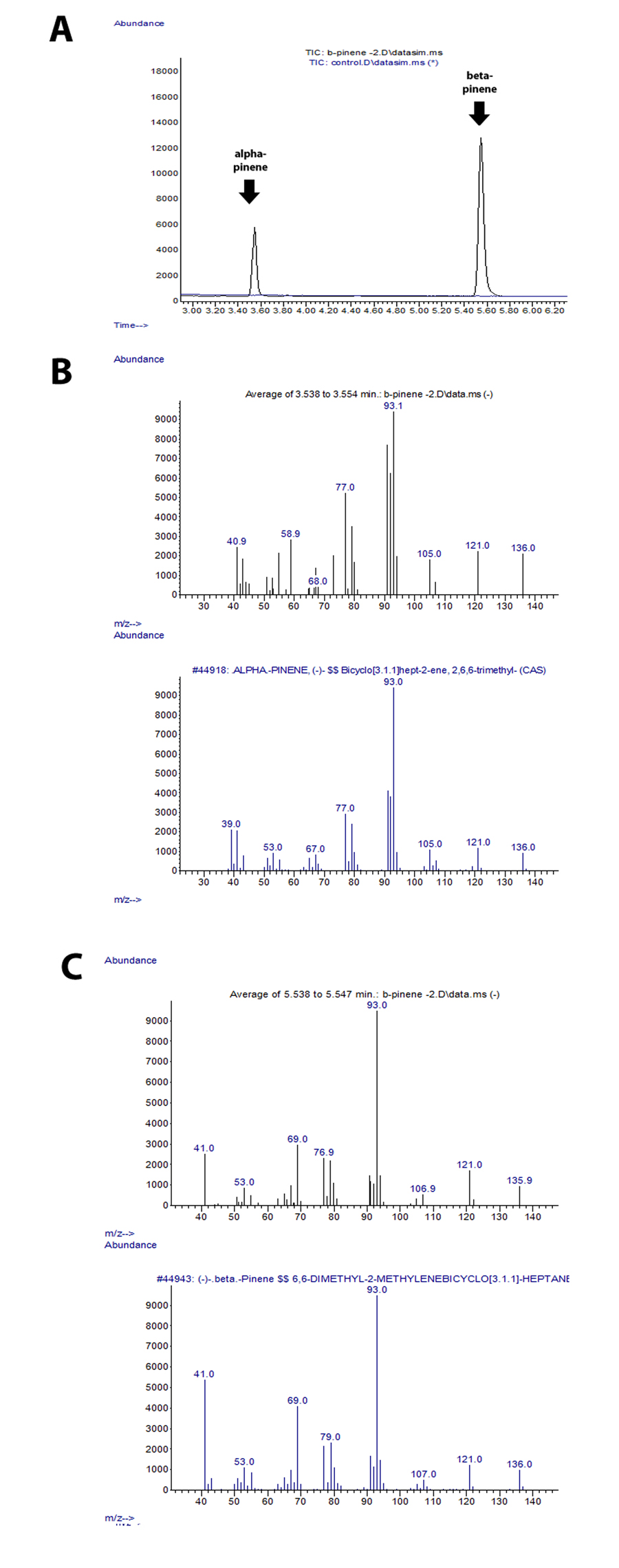
Expression of Beta-pinene bacteria
Daisy purified the pellets of beta-pinene that Joe prepare a week ago and store at -80C. She is certain that there is beta-pinene in these cells because previous SDS-page data shows a strong band slightly below 70kDa ( the size of both alpha and beta-pinene synthases). Well, she performed a GPP assay and prepared those samples for GC-MS , and surprising it came out that the beta-pinene synthase was producing alpha-pinene and mostly beta-pinene:
 "
"