Team:Bielefeld-Germany/Project
From 2011.igem.org
(→References) |
(→References) |
||
| Line 53: | Line 53: | ||
Bi J, Wang H, Xiel J (2011) Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets, [http://onlinelibrary.wiley.com/doi/10.1002/jcp.22419/full ''Journal of Cellular Physiology'' 226(2): 331-340]. | Bi J, Wang H, Xiel J (2011) Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets, [http://onlinelibrary.wiley.com/doi/10.1002/jcp.22419/full ''Journal of Cellular Physiology'' 226(2): 331-340]. | ||
| - | Ghosh S, | + | Ghosh S, George S, Roy U, Ramachandran D, Kolthur-Seetharam U (2010) NAD: A master regulator of transcription [http://www.sciencedirect.com/science/article/pii/S1874939910001057 ''Biochimica et Biophysica Acta'' 1799(10-12): 681-693]. |
Grahnert A, Grahnert A, Klein C, Schilling E, Wehrhahn J, Hauschildt S (2011) Review: NAD<sup>+</sup> : A modulator of immune functions, [http://ini.sagepub.com/content/17/2/212.abstract ''Innate Immunity'' 17(2): 212-233]. | Grahnert A, Grahnert A, Klein C, Schilling E, Wehrhahn J, Hauschildt S (2011) Review: NAD<sup>+</sup> : A modulator of immune functions, [http://ini.sagepub.com/content/17/2/212.abstract ''Innate Immunity'' 17(2): 212-233]. | ||
Revision as of 16:05, 6 September 2011

Contents |
Project description
The development of sensitive and selective Biosensors is an important topic in synthetic biology. Biosensors can be applied in a wide range - from the detection of environmental toxics up to clinical diagnostics. Because cells have to sense their surroundings, there are a lot of natural systems that are similar to a biosensor. Prejudicial cellular biosensors often show negative side effects that complicate any practical application. Common problems are the limited use outside a gene laboratory due to the use of genetically engineered cells, the low durability because of the usage of living cells and the appearance of undesired signals induced by endogenous metabolic pathways.
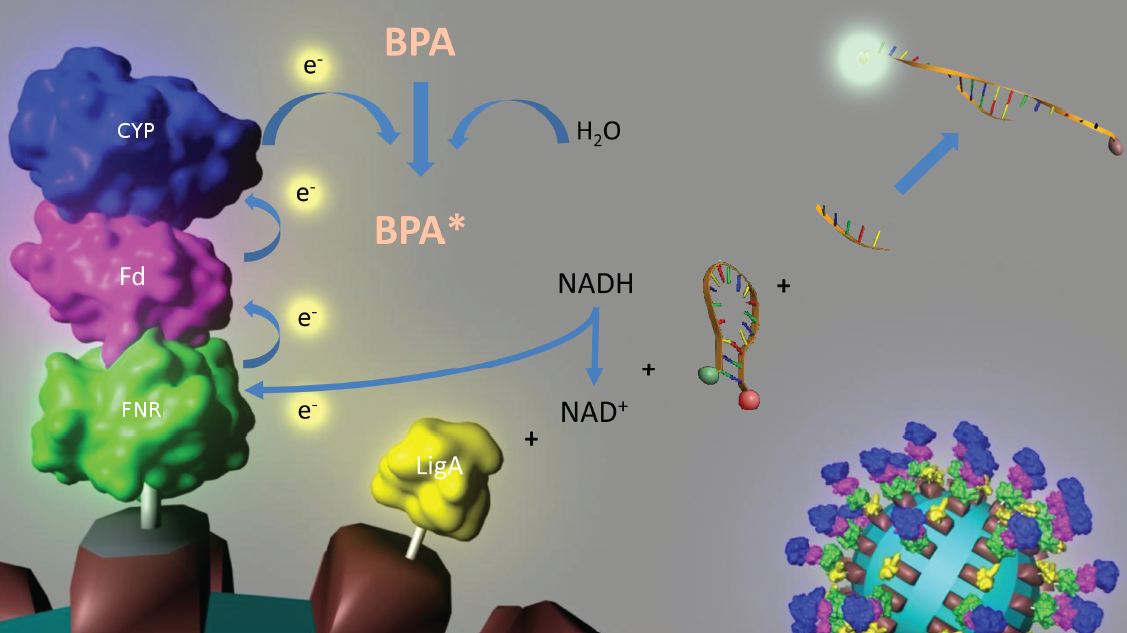
To solve these problems, the iGEM-Team Bielefeld 2011 aims at developing a cell-free bisphenol A (BPA) biosensor based on a coupled enzyme reaction fused to S-layer proteins for everyday use. Bisphenol A is a supposedly harmful substance which is used in the production of polycarbonate. To detect BPA it is degraded by a fusion protein under formation of NAD+ which is detected by an NAD+ dependent enzymatic reaction with a molecular beacon. Both enzymes are fused to S-layer proteins which build up well-defined nanosurfaces and are attached to the surface of beads. By providing these nanobiotechnological building blocks the system is expandable to other applications.
An overview of our project is shown in the figure below. The background and the state of the art of each subproject is described below this figure. To have a quick insight of what is happining in our lab take a look in our Labjournal.
S-layer
S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self assembly-domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to eachother (determined by the S-layer geometry) it is possible to realize various practical applications (Sleytr et al., 2007). The usability of such well defined nano-lattice structures is far-reaching from ultrafiltration membranes to the development of immobilized biosensors.
Especially for the production of cell-free biosensors, functional fusion proteins are of great importance. Kainz et al. (2010) fused fluorescent proteins with an S-layer glycoprotein from Geobacillus stearothermophilus. They demonstrated that the properties of the fusion protein were similar to the native fluorescent protein. The intensity of the fluorescence, the lifetime and the adsorption spectra showed comparable behavior at different pH-values. Enzymes fused to immobilized S-layers showed a significantly longer durability (Schäffer et al., 2007).
The iGEM-Team Bielefeld aims at the assembly, production and immobilization of S-layer fusion proteins for the detection of BPA by a coupled enzymatic reaction. S-layers from five different organisms are employed. The provision of various S-layers with different geometries offers the possibility for the scientific community to create functional nanobiotechnological surfaces with simple and standardized methods, quasi do it yourself nanobiotechnology. First, different fusion proteins with fluorescent proteins and a luciferase are created. The functionality and efficiency of the immobilization to various materials such as silicon dioxide or cellulose is then characterized by measuring the fluorescence and luminescence, respectively.
Bisphenol A degradation
In 2005, Sasaki et al. isolated a soil bacterium from the Sphingomonas genus which is able to degrade the environmental poison bisphenol A (BPA) with a unique rate and efficiency compared to other BPA degrading organisms. This strain was called Sphingomonas bisphenolicum AO1 and is able to completely decompose 120 mg BPA L-1 in about 6 hours. Three genes which are responsible for the first step of this effective BPA degradation by S. bisphenolicum AO1 were identified: a cytochrome P450 (bisdB), a ferredoxin (bisdA) and a ferredoxin-NAD+ oxidoreductase (FNR) (Sasaki et al., 2005b). The bisdAB genes from S. bisphenolicum AO1 were isolated, transformed into and expressed in E. coli and enabled this bacterium to degrade BPA, too (Sasaki et al., 2008). In addition, the BisdAB proteins from S. bisphenolicum AO1 were able to degrade BPA in a cell free system in which spinach reductase was added (Sasaki et al., 2005b). So we assume that the BisdAB proteins also work in a cell free system together with the ferredoxin-NAD(P)+ oxidoreductase from E. coli. The suggested reaction mechanism of the first BPA degradation step is shown in the project overview image above.
In 2008, the iGEM team from the University of Alberta submitted the codon usage optimized bisdAB genes from S. bisphenolicum AO1 to the registry of standard biological parts in the so called Freiburg BioBrick assembly standard (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>). Via this assembly standard it is very easy to build fusion proteins. These already existing protein domains will be fused together with the NAD(P)+ oxidoreductase gene from E. coli to the fusion protein FNR:Fdbisd:P450bisd which subsequently will be fused to an S-layer gene. We have already shown that the Fdbisd:P450bisd fusion protein is degrading BPA more effective in E. coli than the polycistronic bisdAB gene (data here).
NAD+ detection
Our selected NAD+ detection method displays a molecular beacon based approach. These have been initially described in 1996 as nucleic acid probes that fluoresce upon hybridization (Tyagi et al., 1996). For this effect the ends of a single-stranded DNA molecule are labeled with a fluorophore as well as with an appropriate quencher. Both are in close proximity to each other due to a formed stem-loop, so that the detection of any fluorescence signal is prevented.
The molecular beacon’s closed state can be applied to a bioassay detecting NAD+ even in very low concentrations (Tang et al., 2011). Using two complementary targets hybridizing side-by-side with the hairpin enables NAD+-dependent DNA ligation by Escherichia coli DNA ligase. Only after closing the gap between both hybridized targets the stem melts and the secondary structure gets broken down to a linearized probe-target hybrid. The immediate consequence is a disruption of the close proximity of the fluorophore and the quencher, so that an excitation with light is converted into a visible fluorescence signal. Hence, NAD+ concentration determines DNA ligase activity, which is responsible for the formation of the molecular beacon’s open state and therefore directly correlates with the emerging fluorescence signal. Additionally, the highly selective bioassay has a low limit of detection (0,3 nM) compared to other methods.
Because of the signal’s stability and the suitability for daily use the NAD+ bioassay can be coupled to NADH-dependent BPA degradation in the context of biosensing.
Further applications
NAD+ is ubiquitous and an indispensable cofactor for all living organisms (Bi et al., 2011). The molecule has a wide range of physiological functions in cellular metabolism (Fig. 1) and has a great impact on the cell integrity (Grahnert et al., 2011). It is entrenched to the energy metabolism and determines the redox status of the cell due to its purpose as reduction equivalent. This is strongly linked to the control of signaling and transcriptional events. For instance, NAD+ has been described as a modulator of immune functions (Grahnert et al., 2011) and a master regulator of transcription (Ghosh et al., 2010). Several enzymes are tightly regulated by the cellular balance between the oxidized and reduced form of NAD+ and therefore regarded as a potential target for therapeutics. Indeed, NAD+ acts as a substrate for a wide range of proteins including NAD+-dependent protein deacetylases, poly(ADP-ribose) polymerases and several transcription factors (Houtcooper et al., 2010). A disorder of cellular NAD+ level has usually relevance in symptoms and diseases like stroke, cardiac ischemia, epilepsy, huntington disease, wallerian degeneration, cancer, type two diabetes mellitus (T2DM), neutrophil survival, longevity, obesity as well as cardiovascular and neurodegenerative disease (Sauve, 2007, Houtcooper et al., 2010). The regulation of NAD+-dependent pathways may have a major contribution to oxidative metabolism and life span extension (Houtcooper et al., 2010). The biosynthetic pathways for NAD+ are proposed as promising novel antibiotics targets against pathogens (Bi et al., 2011).

The reviewed molecular beacon based NAD+ bioassay can be applied to biochemical and biomedical studies (Tang et al., 2011). Accordingly, it can be utilized to detect NAD+/NADH-dependent enzymatic processes. The low limit of detection, its reliability and manageability provides a practical alternative to present colorimetric, fluorometric, chemiluminscent, electrochemical or mass spectrometric methods detecting NAD+ or NADH. In context of clinical applications the NAD+ bioassay can be useful to monitor cellular NAD+ level during defined treatments of tissues with drug targets, for instance. Finally, the principle of the proposed NAD+ bioassay can be used for optical biosensors by either immobilization of molecular beacons on the sensor surface or their presence directly in the reaction medium (Tang et al., 2011).
References
Bi J, Wang H, Xiel J (2011) Comparative genomics of NAD(P) biosynthesis and novel antibiotic drug targets, Journal of Cellular Physiology 226(2): 331-340.
Ghosh S, George S, Roy U, Ramachandran D, Kolthur-Seetharam U (2010) NAD: A master regulator of transcription Biochimica et Biophysica Acta 1799(10-12): 681-693.
Grahnert A, Grahnert A, Klein C, Schilling E, Wehrhahn J, Hauschildt S (2011) Review: NAD+ : A modulator of immune functions, Innate Immunity 17(2): 212-233.
Houtkooper RH, Cantó C, Wanders RJ, Auwerx J (2010) The Secret Life of NAD+: An Old Metabolite Controlling New Metabolic Signaling Pathways, Endocrine Reviews 31 (2): 194.
Kainz B, Steiner K, Möller M, Pum D, Schäffer C, Sleytr UB, Toca-Herrera JL (2010) Absorption, Steady-State Fluorescence, Fluorescence Lifetime, and 2D Self-Assembly Properties of Engineered Fluorescent S-Layer Fusion Proteins of Geobacillus stearothermophilus NRS 2004/3a, Biomacromolecules 11(1):207-214.
Sasaki M, Maki J, Oshiman K, Matsumura Y, Tsuchido T (2005a) Biodegradation of bisphenol A by cells and cell lysate from Sphingomonas sp. strain AO1, Biodegradation 16(5):449-459.
Sasaki M, Akahira A, Oshiman K, Tsuchido T, Matsumura Y (2005b) Purification of Cytochrome P450 and Ferredoxin, Involved in Bisphenol A Degradation, from Sphingomonas sp. Strain AO1, Appl Environ Microbiol 71(12):8024-8030.
Sasaki M, Tsuchido T, Matsumura Y (2008) Molecular cloning and characterization of cytochrome P450 and ferredoxin genes involved in bisphenol A degradation in Sphingomonas bisphenolicum strain AO1, J Appl Microbiol 105(4):1158-1169.
Sauve AA (2008) NAD+ and Vitamin B3: From Metabolism to Therapies, The Journal of Pharmacology and Experimental Therapeutics 324(3): 883-893.
Schäffer C, Novotny R, Küpcü R, Zayni S, Scheberl A, Friedmann J, Sleytr UB, Messner P (2007) Novel Biocatalysts Based on S-Layer Self-Assembly of Geobacillus Stearothermophilus NRS 2004/3a: A Nanobiotechnological Approach, Small 3(9):1549-1559.
Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, FEMS Microbiol Lett 267(2):131-144.
Tang Z, Liu P, Ma C, Yang X, Wang K, Tan W, Lv X (2011) Molecular Beacon Based Bioassay for Highly Sensitive and Selective Detection of Nicotinamide Adenine Dinucleotide and the Activity of Alanine Aminotransferase, Anal Chem 83(7):2505-2510.
Tyagi S, Kramer FR (1996) Molecular beacons: probes hat fluoresce upon hybridization, Nature Biotechnology 14:303-308.
 "
"