Data Page
Data For Our Favorite New Parts |
BBa_K516210 |
BBa_K516210 (pTet-RBS30-LuxI) - HSL synthesis device, aTc inducible.
This is one of four systems with different RBSs (output modulation) tested to quantify the produced HSL. The results obtained by testing this part allowed the identification of important model parameters to predict the behavior of the whole CTRL+E circuit.
BBa_K516334
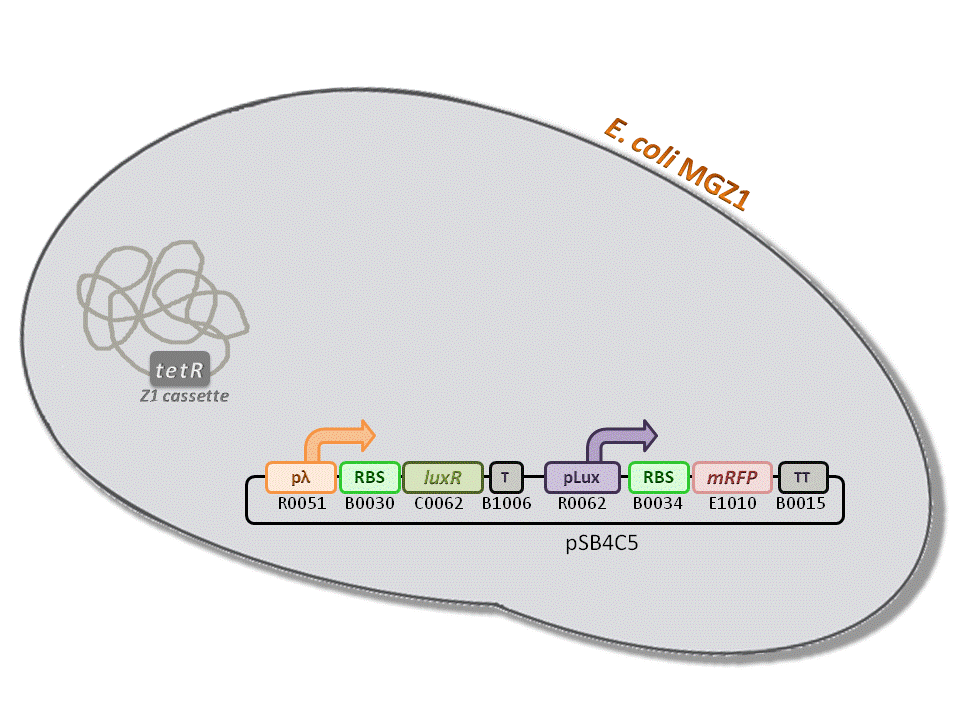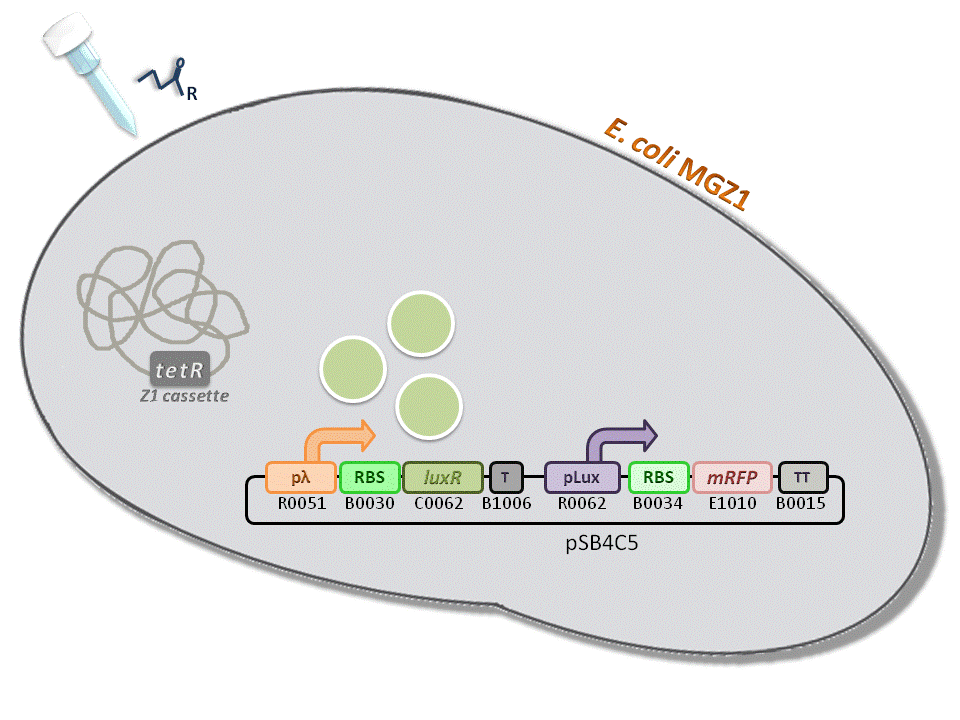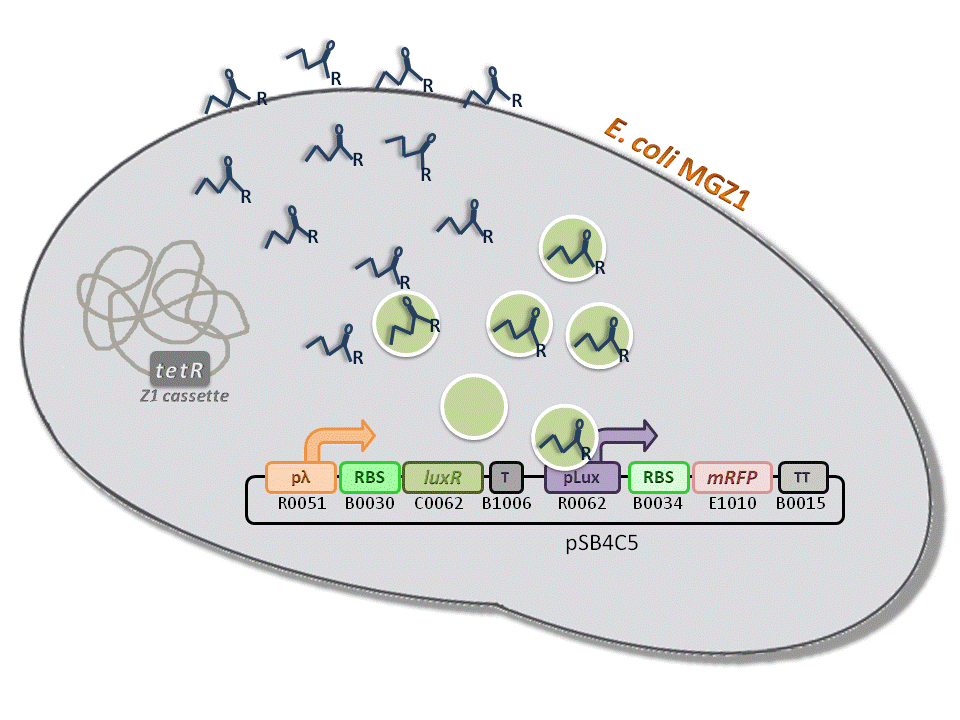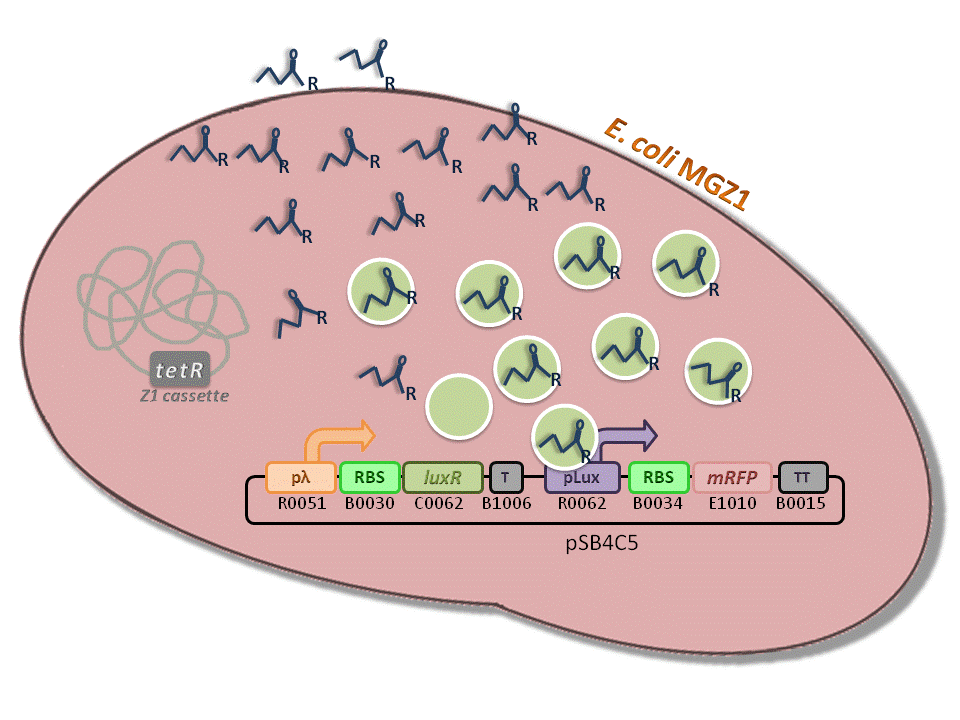
BBa_K516334 (pLambda-RBS30-LuxR-T-pLux-RBS34-mRFP-TT) - 3OC6-HSL biosensor, mRFP output.
This is one of the four biosensors (different RBSs) built and characterized. This measurement system senses the HSL concentration in the medium and responds with an RFP output. Several RBSs have been used to modulate the output range. This is a simple measurement system, used to identify important model parameters used to simulate the behavior of the whole circuit.
BBa_K516230
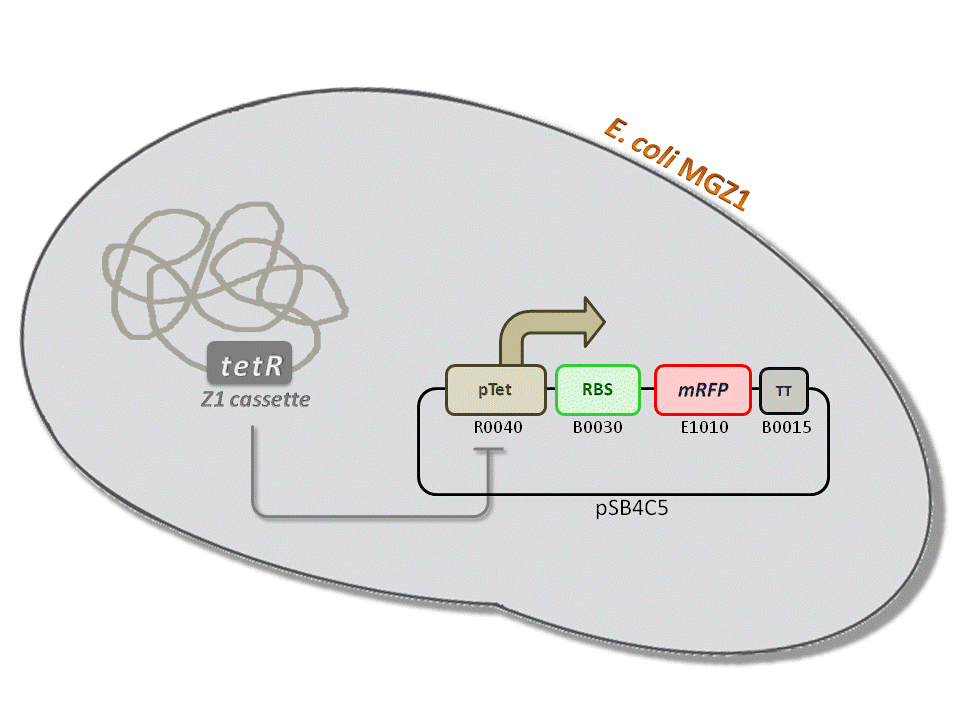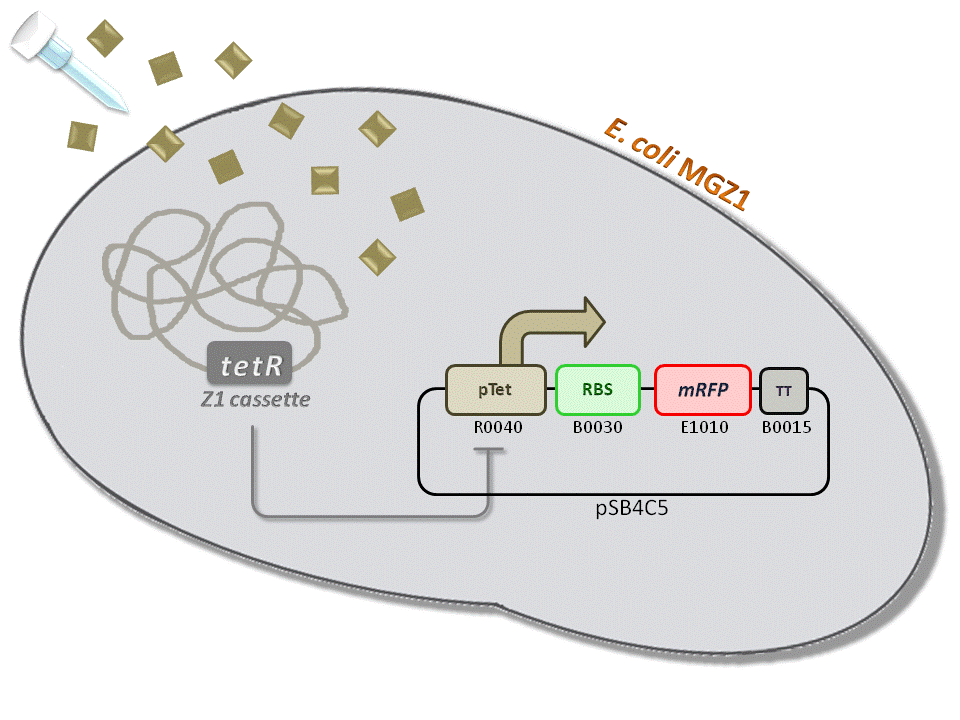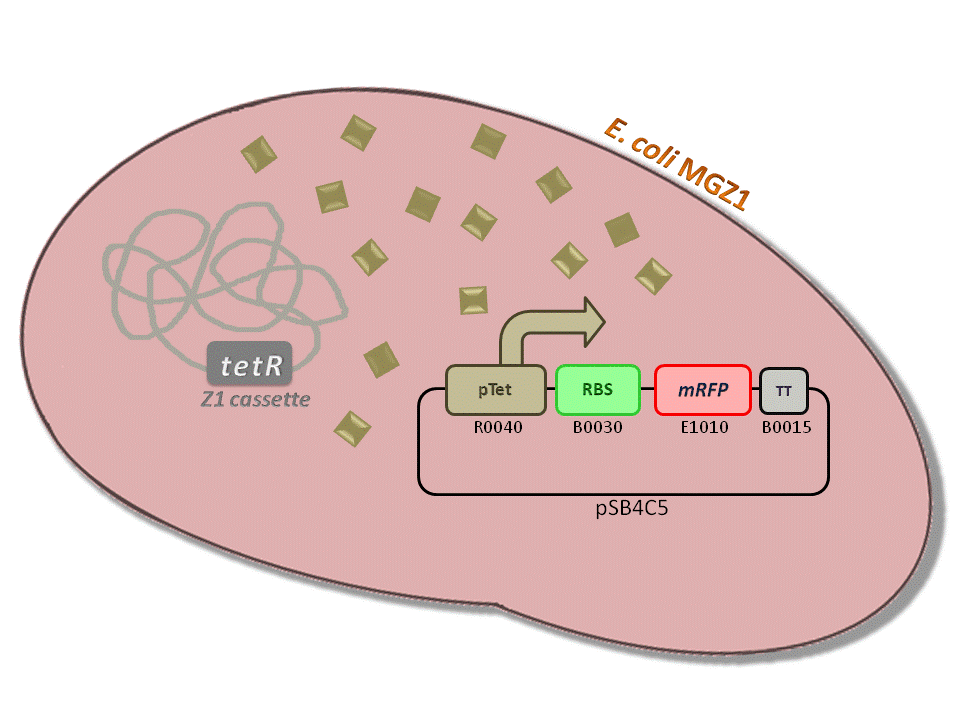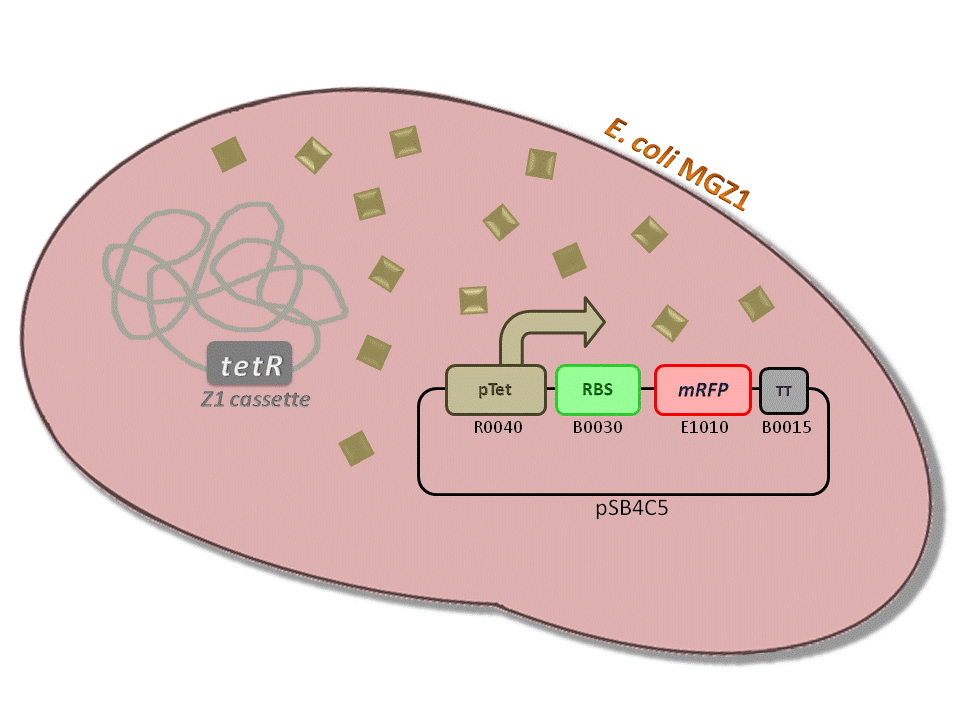
BBa_K516230 (pTet-RBS34-mRFP-TT) - aTc inducible, mRFP output.
This is one of the four pTet measurement systems (different RBSs) built and characterized. This measurement system senses the aTc concentration in the medium and responds with an RFP output. Several RBSs have been used to modulate the output range. This is a simple measurement system, used to identify important model parameters used to simulate the behavior of the whole circuit.
Data For existing Parts
RBSs from the community collection
RBSs (based on Ron Weiss thesis) were used for the fine tuning of CTRL+E. Different experimental conditions were assayed.
Estimated efficiencies in pSB4C5 plasmid with -RBSx-mRFP-TT coding sequence under the control of the specified promoter:
| RBS | effpLux | effpTet | effJ23101 | Declared efficiency |
| B0030 | 0.40 | 1.6814 | 2.45 | 0,6 |
| B0031 | 0.01 | ND | 0.04 | 0,07 |
| B0032 | 0.19 | 0.4193 | 0.40 | 0,3 |
| B0034 | 1 | 1 | 1 | 1 |
Estimated efficiencies in pSB4C5 plasmid with pTet-RBSx-GeneX-TT, with GeneX=mRFP, AiiA or LuxI:
| RBS | effmRFP | effAiiA | effLuxI | Declared efficiency |
| B0030 | 1.72 | 0.53 | 0.45 | 0,6 |
| B0031 | 0.03 | 0.83 | 0.028 | 0,07 |
| B0032 | 0.37 | 0.50 | N.D. | 0.3 |
| B0034 | 1 | 1 | 1 | 1 |
BBa_K081022
- Experience page of BBa_K081022 Plambda regulated luxR generator and Plux.
This device has been characterized with several RBSs in order to modulate the output range. In this way, this device can be used to tightly control the expression of downstream encoded protein from a very weak production (using weak RBS) to a very intensive production (with strong RBSs).
The Hill's estimated parameters are reported in the table below:
| RBS | αpLux [(AUr/min)/cell] | δpLux [-] | ηpLux [-] | kpLux [ng/ml] |
| BBa_B0030 | 438 [10] | 0.05 [>100] | 2 [47] | 1.88 [27] |
| BBa_B0031 | 9.8 [7] | 0.11 [57] | 1.2 [29] | 1.5 [26] |
| BBa_B0032 | 206 [3] | 0 [>>100] | 1.36 [10] | 1.87 [9] |
| BBa_B0034 | 1105 [6] | 0.02 [>100] | 1.33 [19] | 2.34 [18] |
The operative parameters are reported in the table below:
| RBS | RPUmax | RPUmin | Switch point [nM] | Linear boundaries [MIN; MAX] [nM] |
| B0030 | 4.28 | 0.20 | 1.08 | [0.36; 3.27] |
| B0031 | 4.93 | 0.55 | 0.25 | [0.03; 2.30] |
| B0032 | 9.49 | 0.02 | 0.47 | [0.07; 3.07] |
| B0034 | 21.53 | 0.51 | 0.53 | [0.08; 3.77] |
BBa_R0040
- Experience page of BBa_R0040 TetR repressible promoter.
pTet promoter was characterized with various RBSs and mRFP.
The Hill's estimated parameters are reported in the table below:
| RBS | αpTet [(AUr/min)/cell] | δpTet [-] | ηpTet [-] | kpTet [nM] |
| BBa_B0030 | 230.67 [3.7] | 0.028 [91.61] | 4.61 [23.73] | 8.75 [4.16] |
| BBa_B0031 | ND | ND | ND | ND |
| BBa_B0032 | 55.77 [12] | 1.53E-11 [>>100] | 4.98 [57.62] | 7.26 [14.98] |
| BBa_B0034 | 120 [5.95] | 0.085 [40.6] | 24.85 [47.6] | 9 [5.43] |
The operative parameters are reported in the table below:
| RBS | RPUmax | RPUmin | Switch point [ng/ml] | Linear boundaries [MIN; MAX] [ng/ml] |
| B0030 | 1.53 | ~0 | 7.95 | [4.66;11.99] |
| B0031 | ND | ND | ND | ND |
| B0032 | 3.16 | ~0 | 6.7 | [4.45;10.05] |
| B0034 | 2.73 | 0.23 | 8.96 | [8.27;9.71] |
Improved existing parts
BBa_K516999

This vector was designed and realized in order to facilitate the cloning of coding sequences downstream of the strong promoter pTet.
 "
"