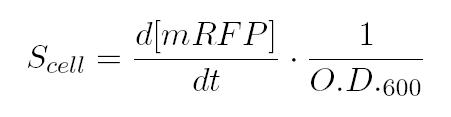CTRL + E
Signalling is nothing without control...
Contents |
Mathematical modelling page
Mathematical modelling plays nowadays a central role in Synthetic Biology, due to its ability to serve as a crucial link between the concept and realization of a biological circuit.
According to this, after a brief overview about the advantages that modelling engineered circuits can bring, we deeply analyze the system of equation formulas, underlining the role and the function of the parameters involved.
Then, experimental procedures for parameters estimation are presented and, finally, different types of circuit are discussed and their simulations performed, using ODE's with MATLAB and explainig the difference between a closed-loop model and an open one.
Then, experimental procedures for parameters estimation are presented and, finally, different types of circuit are discussed and their simulations performed, using ODE's with MATLAB and explainig the difference between a closed-loop model and an open one.
The importance of the mathematical model
Several motivations are strong enough to accept the idea that mathematical model is very useful in this study.
- Firstly in the initial steps of the project, beacuse of its capability to predict the kinetics of the enzymes (aiiA, Luxi) and HSL involved in our gene network, well realizing the a-priori identification in silico in order to understand if the complex circuit's structure and functioning could be achievable.
- Secondly, for the parametric identification. Using the lsqnonlin function of MATLAB it was possible to get all the parameters involved in the model and consequently to know, for example, the shape of the activation curve of the promoters (Plux, Ptet), according to the a-posteriori identification.
- Thirdly, the reproducibility. Studing and characterizing simple subparts can allow us not only to predict the behavior of the final circuit, but also it can be useful in other studies, facing with the same basic modules.
Equations for gene networks
Equations (1) and (2)
We have condensed in a unique equation transcription and translation processes. Equations (1) and (2) have identical structure, differing only in the parameters involved.
The first term describes, through Hill's equation formalism, the synthesis rate of the protein of interest (either LuxI or AiiA) depending on the concentration of the inducible protein (anhydrotetracicline -aTc- or HSL respectively). As can be seen in the parameters table (see below),α refers to the maximum activation of the promoter, δ stands for its leakage activity (this means that the promoter is quite induced even if there is no input). In particular, in equation (1), the quite total inhibition of pTet promoter is due to the constitutive production of TetR by our MGZ1 strain, while, in equation (2), Plux is almost repressed in the absence of the complex given by LuxR and HSL.
In the first term of equation (2) we have described the inducer as being represented only by HSL. This formalism stems from the fact that our final device offers a constitutive production of LuxR (due to the upstream constitutive promoter pLac), so that, assuming it abundant in the cytoplasm, we can derive the semplification of attributing pLux promoter induction only by HSL: this is the reason why we didn' t consider LuxR in the equations system as well as LuxI and AiiA.
Furthermore, in both equations (1) and (2) k and η stands respectively for the other parameter of the Hill relationship. The second term in equation (1) and (2) is composed of two parts. The first one (γ*LuxI/AiiA) describes with a linear relation the degradation rate per cell of the protein. The second one (μ*(Nmax-N)/Nmax)*LuxI/AiiA) takes into account the dilution term and is related to the cell replication process.
In the first term of equation (2) we have described the inducer as being represented only by HSL. This formalism stems from the fact that our final device offers a constitutive production of LuxR (due to the upstream constitutive promoter pLac), so that, assuming it abundant in the cytoplasm, we can derive the semplification of attributing pLux promoter induction only by HSL: this is the reason why we didn' t consider LuxR in the equations system as well as LuxI and AiiA.
Furthermore, in both equations (1) and (2) k and η stands respectively for the other parameter of the Hill relationship. The second term in equation (1) and (2) is composed of two parts. The first one (γ*LuxI/AiiA) describes with a linear relation the degradation rate per cell of the protein. The second one (μ*(Nmax-N)/Nmax)*LuxI/AiiA) takes into account the dilution term and is related to the cell replication process.
Equation (3)
The processes described here are not those of transcription and translation, but in principle are enzymatic reactions either related to the production or the degradation of HSL. Based on the experiments performed, we derived Hill's equation in the case of η=1. They cannot be exactly defined Michaelis Menten's equations since that in our formalism, LuxI and AiiA aren't described as enzymes (since they appear also in the denominator). We simply derived empirical formulas relating either LuxI or AiiA to HSL, and treated them with the typical Michaelis Menten formalism since they presented the corresponding sigmoidal shape/switching like behaviour. Regarding to this, we believe that the saturation phenomenon observed either in HSL production rate due to LuxI, or HSL degradation rate due to AiiA, underlies limiting elements in cell metabolism.
In the cell HSL binds to LuxR, and two HSL molecules form a tetramer with two complementary LuxR molecules to form a complex which can then activate the promoter Plux.
Intuitively, LuxI activity as an enzyme encounters an intrinsic limit in HSL synthesis depending on the finite and hypothetically fixed substrate concentration (namely SAM and hexanoyl-ACP, see ref.); this means that at a certain LuxI concentration, all the substrate forms activation complexes with LuxI, so that there is no more substrate available for the other LuxI produced. HSL degradation rate is limited by its availability; even if the concentration varies with time, there is always a corresponding limit in AiiA concentration, which determines a saturation in the degradation rate.
Moreover, both the formulas relating either LuxI or AiiA to HSL are multiplied by the number of cells N, due to the property of the lactone to diffuse free inside/outside bacteria. The third term in equation (3) is similar to the corresponding ones present in the first two equations and describes the intrinsic protein degradation.
Intuitively, LuxI activity as an enzyme encounters an intrinsic limit in HSL synthesis depending on the finite and hypothetically fixed substrate concentration (namely SAM and hexanoyl-ACP, see ref.); this means that at a certain LuxI concentration, all the substrate forms activation complexes with LuxI, so that there is no more substrate available for the other LuxI produced. HSL degradation rate is limited by its availability; even if the concentration varies with time, there is always a corresponding limit in AiiA concentration, which determines a saturation in the degradation rate.
Moreover, both the formulas relating either LuxI or AiiA to HSL are multiplied by the number of cells N, due to the property of the lactone to diffuse free inside/outside bacteria. The third term in equation (3) is similar to the corresponding ones present in the first two equations and describes the intrinsic protein degradation.
Equation (4)
Equation (4) is the common equation describing logistic cell growth, depending on the rate μ and the maximum number NMAX of cells per well reachable.
Table of parameters
| Parameter | Description | Unit of Measurement | Value |
| αPTet | maximum transcription rate of Ptet | [(AUr/min)/cell] | - |
| δPTet | leakage factor of promoter Ptet basic activity | [-] | - |
| ηPTet | Hill coefficient of Ptet | [-] | - |
| kPTet | dissociation costant of Ptet ? | [nM] | - |
| αPLux | maximum transcription rate of Plux | [(AUr/min)/cell] | - |
| δPLux | leakage factor of promoter Plux basic activity | [-] | - |
| ηPLux | Hill coefficient of Plux | [-] | - |
| kPLux | dissociation costant of Plux ? | [nM] | - |
| γPLux | LuxI costant degradation | [1/min] | - |
| γAiiA | AiiA costant degradation | [1/min] | - |
| γHSL | HSL costant degradation | [1/min] | - |
| Vmax_LuxI | maximum transcription rate of LuxI | [nM/(min*cell)] | - |
| km_LuxI | dissociation costant ? | [AUr/cell] | - |
| kCAT | ?? | [1/(min*cell)] | - |
| km_AiiA | dissociation costant ? | [AUr/cell] | - |
| NMAX | maximum number of bacteria per well | [cell] | - |
| μ | rate of bacteria groth | [1/min] | - |
According to the table above, the unit of the state variables are:
| State variable | Unit of Measurement |
| d[LuxI]⁄dt | [AUr⁄(min*cell)] |
| d[AiiA]⁄dt | [AUr⁄(min*cell)] |
| d[HSL]⁄dt | [nM⁄(min)] |
| d[N]⁄dt | [cell⁄(min)] |
Parameter estimation
In this section we examine the parameters of the model and justify the units of measure, relating them to the experiments performed for the characterization of the parts. We want to underline again our concept of modelling: beginning to caractherize simplier parts, we get their parameters and we try to predict the behaviour of the final engineerd closed-loop.
Promoter (Ptet & Plux)
These are the first subparts tested. In this phase of the project we aim to increase our knowledge about promoter Ptet and Plux. Keeping this idea in mind, let's talk about the steps to estimate parameters.
Firstly, in the figure above "RBSx" stands for, respectively, RBS30, RBS31, RBS32, RBS34. So we get 4 biobricks for each promoter, in order to investigate what happens in different conditions of RBS's efficiency. Then, under a wide (more or less, depending on the type of test) range of induction (aTc, HSL for Ptet, Plux respectively) we are going to monitor, in time, absorbance and florescence.
After that, we only consider the exponential gap of bacteria' s groth, and here we calculate the Scell as:
Firstly, in the figure above "RBSx" stands for, respectively, RBS30, RBS31, RBS32, RBS34. So we get 4 biobricks for each promoter, in order to investigate what happens in different conditions of RBS's efficiency. Then, under a wide (more or less, depending on the type of test) range of induction (aTc, HSL for Ptet, Plux respectively) we are going to monitor, in time, absorbance and florescence.
After that, we only consider the exponential gap of bacteria' s groth, and here we calculate the Scell as:
 "
"