Team:DTU-Denmark/Project
From 2011.igem.org
Project
Contents |
The abstract
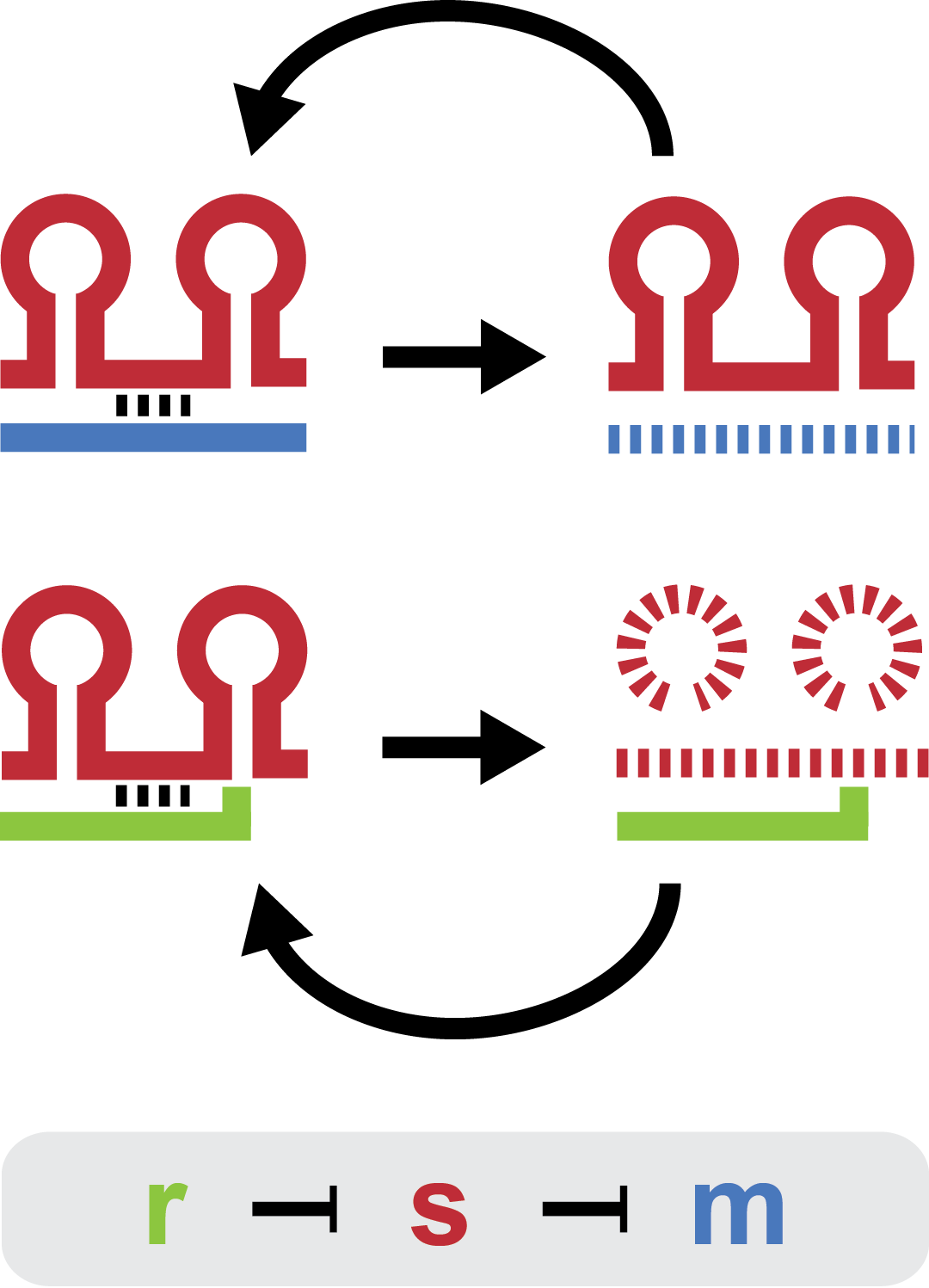
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. A novel type of small RNA regulation displaying favorable properties was investigated ...
Experiments
Verifying that the envisioned small RNA based gene silencing is possible. Plasmids containing and strains deleted for the components were constructed providing a biological model. The dynamic range of the araBAD promoter was expanded. For full description see experiments
Modeling
Modeling provides a framework for characterization and the means to incorporate the trap-RNA system into larger models. A steady state analysis revealed that each trap-RNA system has a characteristic fold repression. The influence of parameters on the fold repression was investigated to help guide the design of the trap-RNA system. For more information got to modeling.
Bioinformatic
A bioinformatic study was performed to investigate the possibilities of engineering the trap-RNA system. The aim of engineering being to target any mRNA and to change properties of the system. The study elucidates interesting features of sequence and secondary structure conservation guiding future genetic engineering. For full analysis go to bioinformatic.
References
[1] Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 97, 6640-6645(2000).
[2] Figueroa-Bossi, Nara, Martina Valentini, Laurette Malleret, and Lionello Bossi. “Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.” Genes & Development 23, no. 17 (2009): 2004 -2015.
[3] Hayashi, K. et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Molecular Systems Biology 2, 2006.0007(2006).
[4] Lambert, J.M., Bongers, R.S. & Kleerebezem, M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Applied and environmental microbiology 73, 1126-35(2007).
[5] Overgaard, Martin, Jesper Johansen, Jakob Møller‐Jensen, and Poul Valentin‐Hansen. “Switching off small RNA regulation with trap‐mRNA.” Molecular Microbiology 73, no. 5 (September 2009): 790-800.
 "
"