Team:DTU-Denmark/Project
From 2011.igem.org
(→Modeling) |
|||
| Line 17: | Line 17: | ||
A bioinformatic study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. For full analysis go to [[Team:DTU-Denmark/Bioinformatic|bioinformatic]]. | A bioinformatic study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. For full analysis go to [[Team:DTU-Denmark/Bioinformatic|bioinformatic]]. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{:Team:DTU-Denmark/Templates/Standard_page_end}} | {{:Team:DTU-Denmark/Templates/Standard_page_end}} | ||
Revision as of 17:55, 20 September 2011
Project
Contents |
The abstract
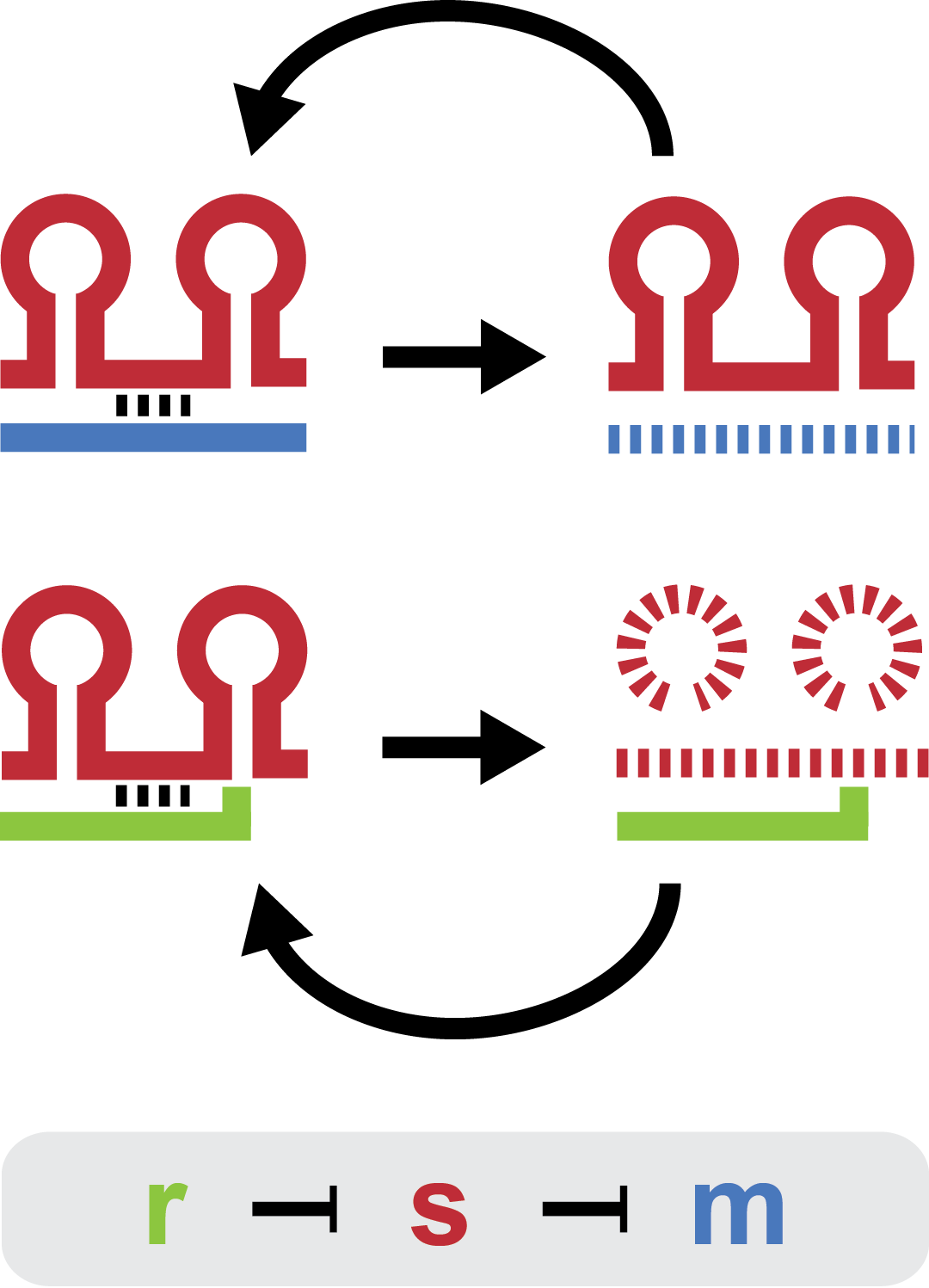
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. A novel type of small RNA regulation displaying favorable properties was investigated ...
Experiments
Verifying that the envisioned small RNA based gene silencing is possible. Plasmids containing and strains deleted for the components were constructed providing a biological model. The dynamic range of the araBAD promoter was expanded. For full description see experiments
Modeling
A framework for characterization was developed to guide rational design and test hypotheses. Steady state analysis revealed that each trap-RNA system has a characteristic fold repression. For more information got to modeling.
Bioinformatic
A bioinformatic study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. For full analysis go to bioinformatic.
 "
"