Team:DTU-Denmark/Project
From 2011.igem.org
(→The models) |
(→The experiments) |
||
| Line 10: | Line 10: | ||
== The experiments == | == The experiments == | ||
| - | The experiments are a proof of concept, showing that the sRNA and the trap-RNA can be used to silence and control gene expression by targeting the chitoporin | + | The experiments are a proof of concept, showing that the sRNA and the trap-RNA can be used to silence and control gene expression by targeting the chitoporin Shine-Dalgarno. The experiments fall into '''three parts'''. For full description go to [[Team:DTU-Denmark/Project_experiment|experiments]]. |
'''Construction of plasmids''' necessary for testing our system involves taking the native system from ''E. coli'', as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system. | '''Construction of plasmids''' necessary for testing our system involves taking the native system from ''E. coli'', as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system. | ||
Revision as of 12:54, 20 September 2011
Project
Contents |
Abstract
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of synthetic biological circuits, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation[2][5], where the inhibition caused by a small regulatory RNA is relieved by another RNA called trap-RNA. The system displays a large dynamic range and can uniquely target and repress any gene of interest providing unprecedented flexibility. We suspect that any level of repression is achievable by simply altering the sequences of the involved RNAs. Multiple such systems can coexist without interfering and are thus compatible with more complex designs.
The natural system
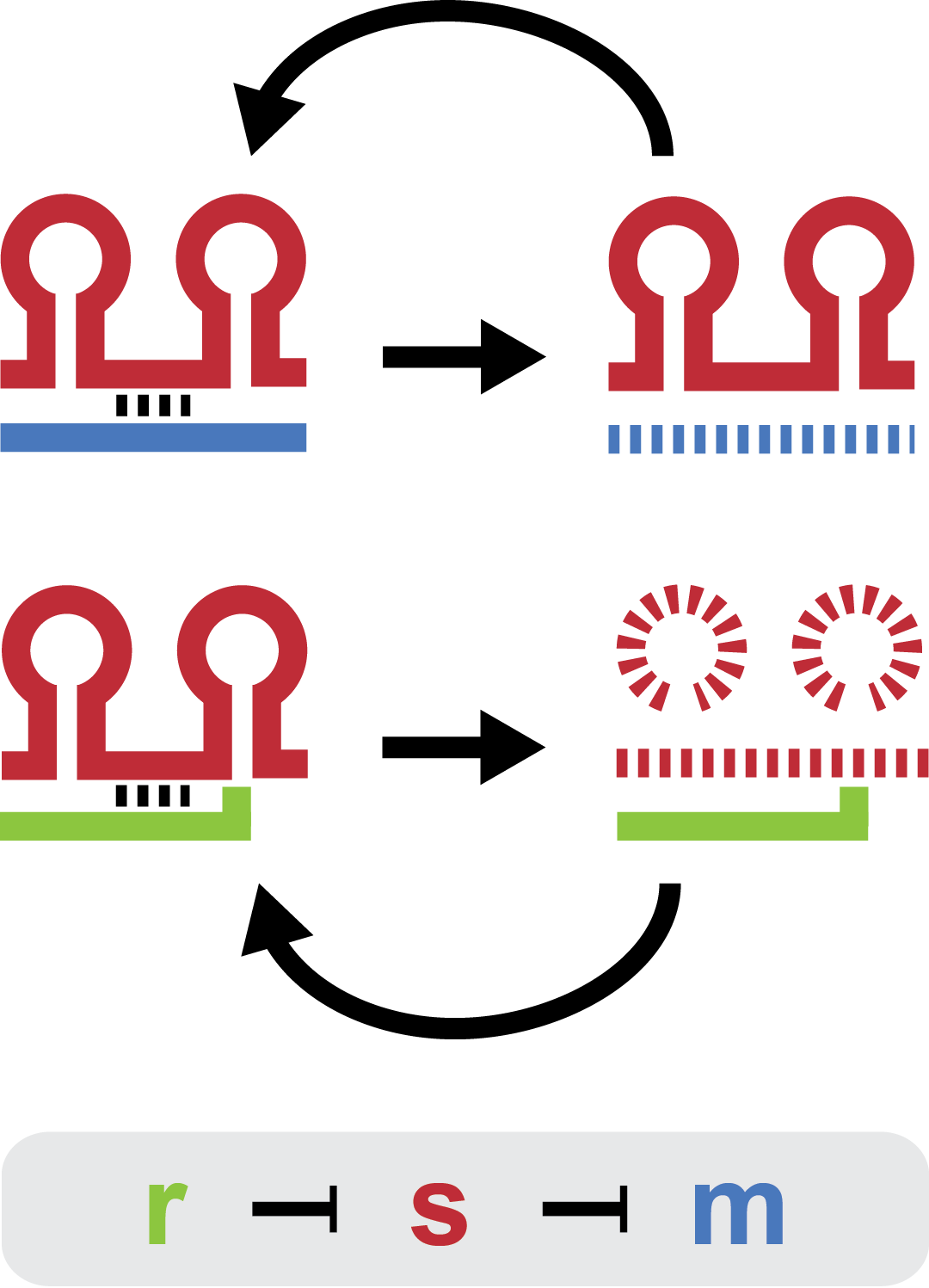
The project is inspired by the regulation of chitobiose uptake and metabolism in E. coli. Especially the three components: a chitoporin that transports chitosugars into cells; a sRNA which regulates the chitoporin post-transcriptionally; and a trap-RNA which is transcribed from intergenic region in the chitobiose operon. The sRNA regulates chitobiose expression through binding to the Shine_Dalgarno sequence on the chitobiose mRNA inhibiting recruitment of the 30S ribosome and altering RNA stability. When chitobiose is present trap-RNA is transcribed and its transcript binds sRNA, relieving chitoporin repression.
The experiments
The experiments are a proof of concept, showing that the sRNA and the trap-RNA can be used to silence and control gene expression by targeting the chitoporin Shine-Dalgarno. The experiments fall into three parts. For full description go to experiments.
Construction of plasmids necessary for testing our system involves taking the native system from E. coli, as well as a slightly modified system and putting them on plasmids that let us both control the expression of these components and measure the output of the system.
Strain construction involves deleting the original genes from the chromosome of a E. coli W3110 strain. Since we use the original genes, these need to be deleted from the chromosome to prevent them from interfering with our measurements.
Improving the araBAD promoter entails expanding the dynamic range of this promoter by modifying the -10 and -35 sequence of the promoter, as well as randomly changing the nucleotide sequence around and in between these sequences. Since the araBAD promoter is used in our project improving this promoter could lead to even finer control of our system.
The models
Modeling provides a framework for characterization and the means to incorporate the system into larger models. A steady state analysis revealed that each trap-RNA system has a characteristic fold repression. The influence of parameters on the fold repression was investigated to help guide the design of the trap-RNA system. Temporal simulation provides additional tools for characterization and design. For more information got to modeling.
The bioinformatics
A bioinformatic study was performed to investigate the flexibility when engineering sRNA regulation genetically. It elucidates sequences of conservation and helps guide the design process. For full analysis go to bioinformatic.
References
[1] Datsenko, K.A. & Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 97, 6640-6645(2000).
[2] Figueroa-Bossi, Nara, Martina Valentini, Laurette Malleret, and Lionello Bossi. “Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.” Genes & Development 23, no. 17 (2009): 2004 -2015.
[3] Hayashi, K. et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Molecular Systems Biology 2, 2006.0007(2006).
[4] Lambert, J.M., Bongers, R.S. & Kleerebezem, M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Applied and environmental microbiology 73, 1126-35(2007).
[5] Overgaard, Martin, Jesper Johansen, Jakob Møller‐Jensen, and Poul Valentin‐Hansen. “Switching off small RNA regulation with trap‐mRNA.” Molecular Microbiology 73, no. 5 (September 2009): 790-800.
 "
"