Team:Bielefeld-Germany/Results
From 2011.igem.org

Contents |
Bisphenol A degradation
Sequencing results
The iGEM team from the University of Alberta sent in BioBricks for BPA degradation in 2008 (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>). Sequencing of these BioBricks by the iGEM HQ and by us led to negative results so the sequences entered into the partsregistry are not correct although these sequences are the ones from Sphingomonas bisphenolicum AO1 (compare [http://www.ncbi.nlm.nih.gov/nuccore/AB255167.1 genbank entry]). In addition, there are "illegal" AgeI and NgoMIV restriction sites in the BioBrick which are used for Freiburg BioBrick assembly standard (RFC 25). After translating and comparing the original sequences from the partsregistry and the sequences from our sequencing results in silico we saw, that the amino acid sequences were identically. These BioBricks were probably synthesized in the Freiburg assembly standard 25 because they have the accordant restriction sites and they were codon optimized for Escherichia coli but the original sequence from S. bisphenolicum AO1 were entered into the registry because amino acid sequence of the real sequence and the sequence that was entered are identical. The alignments are shown in fig. 1 - 4.
Bisphenol A degradation with E. coli
The bisphenol A degradation with the BioBricks <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> works in E. coli KRX in general. Because Sasaki et al. (2008) reported problems with protein folding in E. coli which seem to avoid a complete BPA degradation, we did not use the strong T7 promoter for expressing these BioBricks but a medium strong constitutive promoter (<partinfo>J23110</partinfo>). With this promoter upstream of a polycistronic bisdAB gene we were able to completely degrade 120 mg L-1 BPA in about 30 - 33 h. By fusing <partinfo>K123000</partinfo> and <partinfo>K123001</partinfo> together we could improve the BPA degradation of E. coli even further, so 120 mg L-1 BPA can be degraded in 21 - 24 h. This data is shown in the following figure:
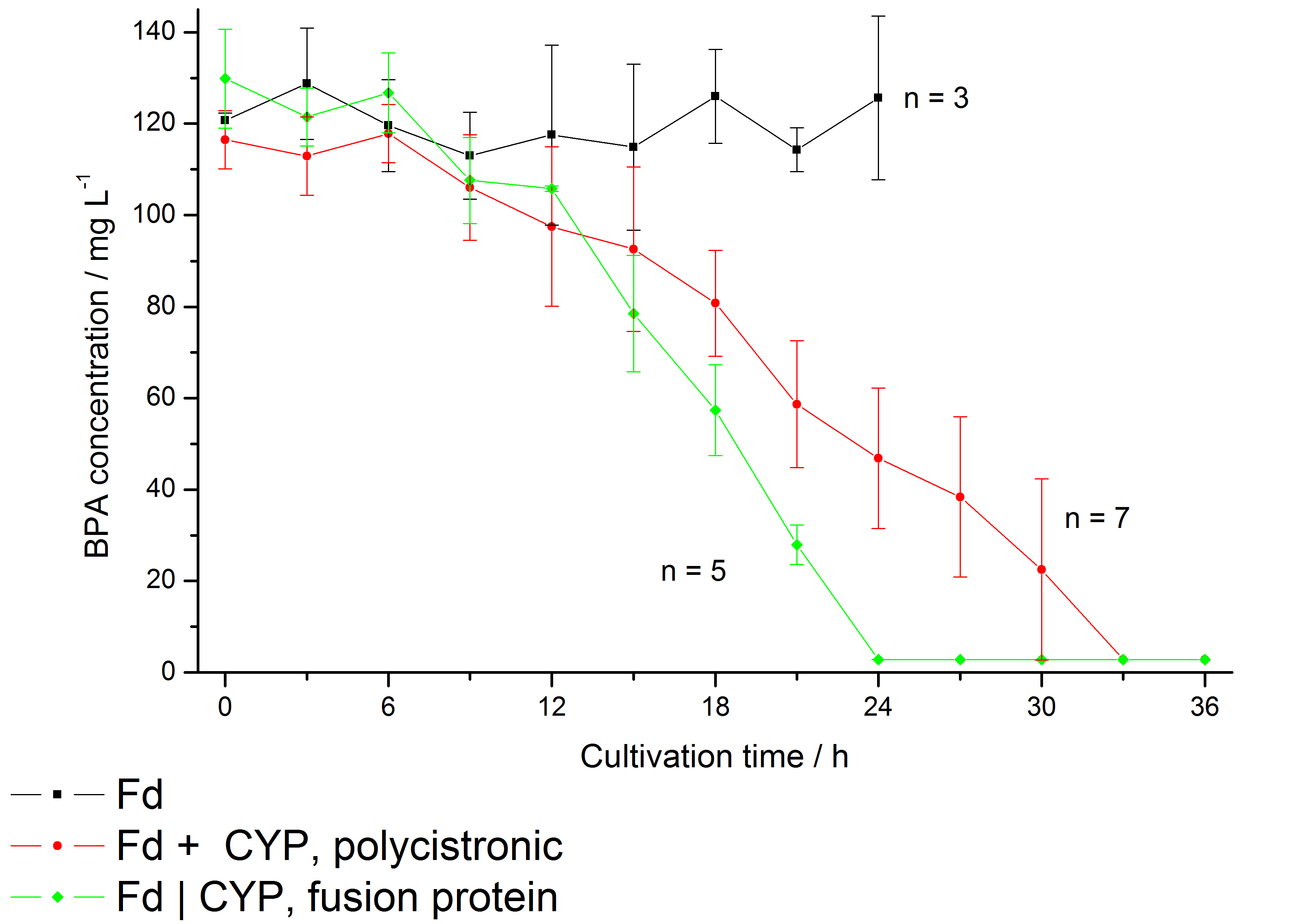
 "
"




