Team:Harvard/Results
From 2011.igem.org
Overview | Biobricks | Source Code | Acknowledgments | Accomplishments
Contents |
Novel Zinc Fingers
Having designed and assembled zinc finger proteins in addition to optimizing our selection system, we were able to test and discover up to 15 novel zinc fingers that potentially binds to the colorblindness gene target.
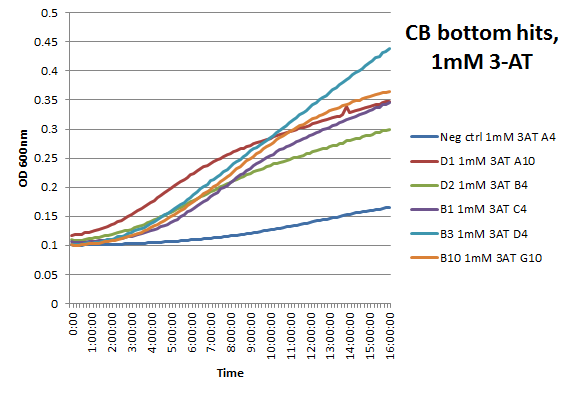
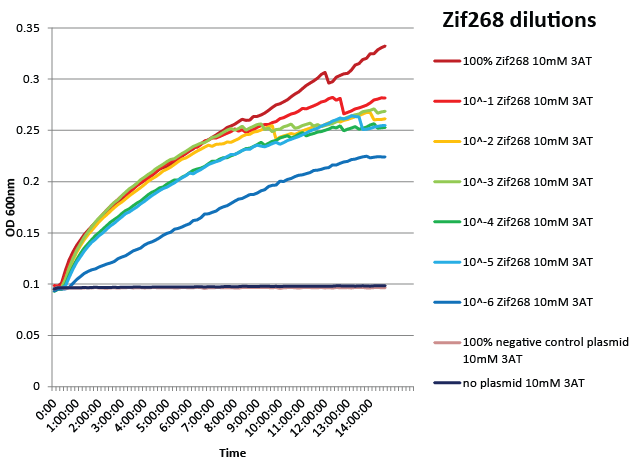
We did not have time to test all 6 targets as planned: we chose to focus on zinc finger binders to DNA tripplet TGG, which is found in the color blindness gene bottom target. We assembled the Color Blindness Bottom library, transformed it into the proper selection strain, and plated on incomplete media with varying concentrations of 3-AT. We saw colonies form, with more on the less stringent plates (NM) and fewer colonies on the higher 3-AT concentrations. Several colonies were picked and tested under varying concentrations of 3-AT in plate reader overnight, and there was a clear difference in growth between the colonies and the negative control (CB bottom binding site with Zif268).
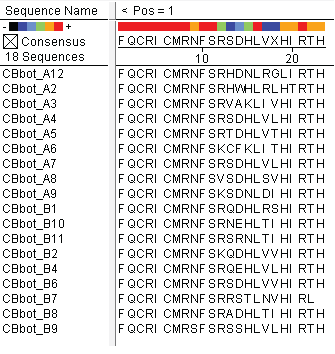
These colonies were then sequenced to discover which of our library zinc fingers was successfully binding to the 5'-GTG GGA TGG-3' sequence. So far, preliminary results indicate that 15 of the colonies have sequences consistent with the DNA sequences on the chip that was submitted, meaning that we have discovered up to 15 novel zinc fingers. The sequences consistent with the chip are CBBot_A1, A3, A4 (same as A7), A6, A8, A9, B1, B10, B11, B2, B4, B6, B7, B8, and B9.
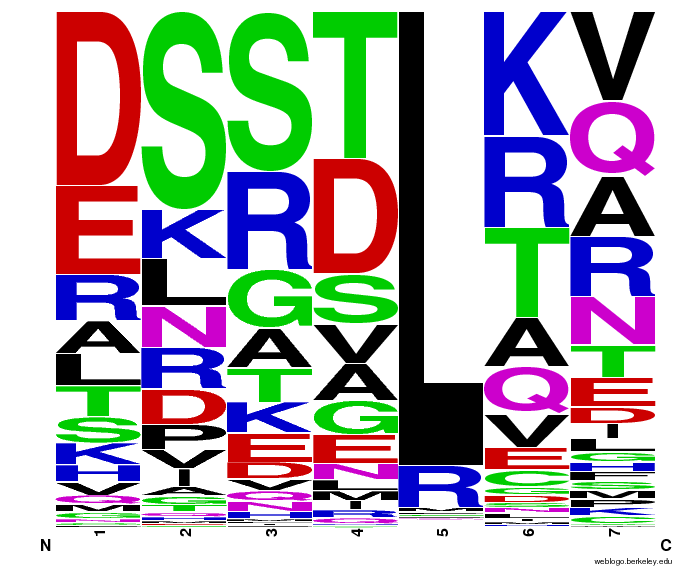
All of the sequenced colonies used the Zif268 backbone, but since the TGG codon is not unlike known Zif268-based zinc finger binding sequences, this may imply that Zif268 is the optimal backbone for such a base triplet. The actual DNA-binding helix is much more variable, and we will continue to test the strength of the binding interactions of these zinc fingers and look for trends in their amino acid make-up.
See our Test page for details on how we used MAGE and lambda red to build and optimize our selection strain.
Genome-Based One-Hybrid Selection Strain
We designed a one-hybrid metabolic system that was entirely genome-based. Using multiplex automated genome engineering (MAGE) and lambda red, we knocked out HisB, PyrF, and rpoZ; inserted a kanamycin cassette-zinc finger binding site-His3-URA3 construct into the 1529620 locus; and changed the zinc finger binding site directly on the genome. The strain was fully characterized and was sensitive enough to recognize a valid zinc finger when diluted as much as one into one million of negative controls. See here for more details.
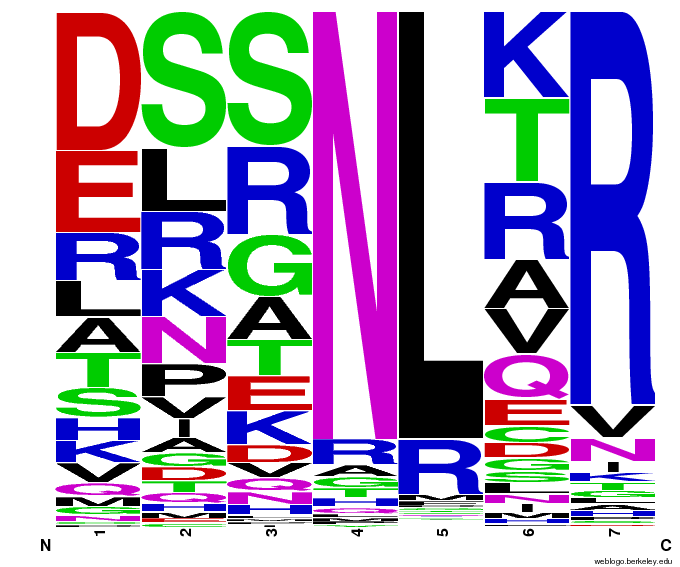
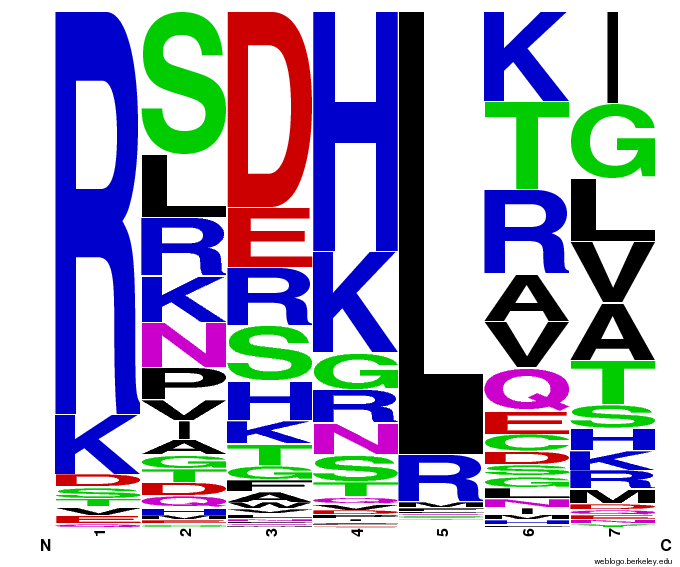
Bioinformatics: 55,000 Potential Zinc Fingers
We made 55,000 sequences, distributed evenly among 6 DNA target triplets. That's 9150 per target.
Because our program's output changes dramatically based on the input triplet, no two sets of sequences are the same:
Make your own zinc finger sequences using our [http://sourceforge.net/projects/harvardigem/ generator]. Written in Python 2.7, this program is what team members created to generate 55,000 zinc finger sequences, as described on our design page.
See our Design page for details on the computational aspects of our project technology.
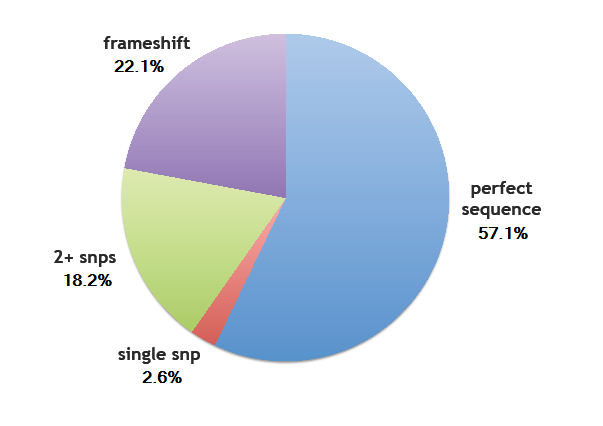
Sequencing Results of Library Transformation
Error rates
- Perfect sequence matches a designed ZF: 57.1% (44/77)
- Single SNP: 2.6% (2/77)
- Two or more SNPs: 18.2% (14/77)
- Frame shift: 22.1% (17/77)
We determined the overall per base pair error rate for this set sequenced to be around 1/200, which includes errors generated by the chip, or generated during PCR and assembly. This is a bit higher than the 1/500 error rate found by Kosuri et al[1], but within a reasonable margin.
SNPs are not as large of a problem as they may seem: they add variety to the library, which is exactly what we originally wanted. While these sequences were not ones that we designed, they may still have produced valid zinc fingers. See Novel Zinc Fingers to see the sequences that are potential hits: while we only considered sequences consistent with our chip (sequences that we designed) to be potentially zinc fingers, the others are also being characterized.
Distributions
Of the 77 samples with good sequencing results, 2 sequences were repeated once. Discounting these, 73 of the 77 sequences, or 94.8%, were unique, showing substantial variability within the library.
See our Synthesize page for details on how we applied chip synthesis to zinc finger proteins.
References
1. Sriram Kosuri, Nikolai Eroshenko, Emily M LeProust, Michael Super, Jeffrey Way, Jin Billy Li, George M Church. (2010). Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nature Biotechnology, 28(12):1295-9. [http://www.nature.com/nbt/journal/v28/n12/full/nbt.1716.html]
 "
"