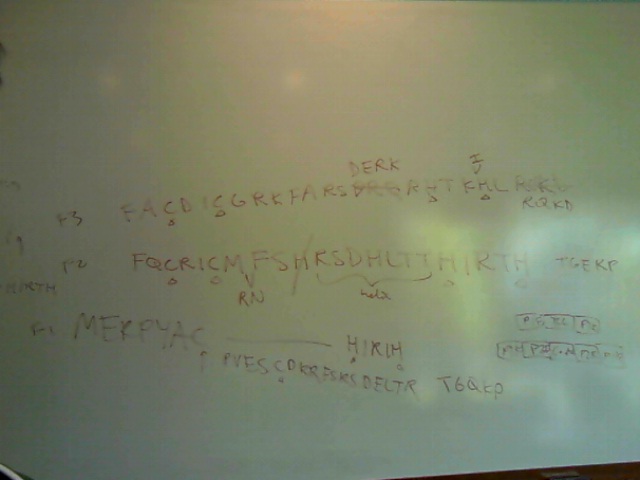Team:Harvard/Notebook
From 2011.igem.org
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| 19 | 20 | 21 | 22 | 23 | 24 | 25 |
| 26 | 27 | 28 | 29 | 30 |
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
| 1 | 2 | |||||
| 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| 10 | 11 | 12 | 13 | 14 | 15 | 16 |
| 17 | 18 | 19 | 20 | 21 | 22 | 23 |
| 24 | 25 | 26 | 27 | 28 | 29 | 30 |
| 31 |
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| 28 | 29 | 30 | 31 |
| Sun | Mon | Tue | Wed | Thu | Fri | Sat | |
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | |||||
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | |
| 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
| 25 | 26 | 27 | 28 | 29 | 30 | 31 |
June 6th
First day of iGEM!June 7th
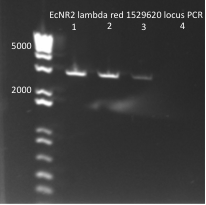
Miniprep of pKD42 (lambda red)
The lambda red plasmid is needed to enable the recombination used to insert the selection/expression systems into our E. coli cultures.
Procedure: followed Qiagen Kit instructions, each student (8) using 1 mL cell suspension
Results: DNA reasonably pure (260/280 between 1.8 and 2) and between 25 and 50 ng/µLJune 8th
PCR to connect ultramers into OZ052 (Zif268 F2 triplicate, GCCGATGTC)and OZ123 (Zif268 F2 triplicate, GAGTGGTTA):
OZ052:
- 3µL OZ052_F (10µM stock)
- 3µL OZ052_R (10µM stock)
- 5µL 10x Pfx amplification buffer
- 1.5µL dNTPs
- 1µL MgSO4
- 0.4µL DNA polymerase
- 36.1µL ddH2O
OZ123:
- 3µL OZ123_F (10µM stock)
- 3µL OZ123_R (10µM stock)
- 5µL 10x Pfx amplification buffer
- 1.5µL dNTPs
- 1µL MgSO4
- 0.4µL DNA polymerase
- 36.1µL ddH2O
Parameters:
- 1) 94⁰C for 5 min
- 2) 94⁰C for 15 sec
- 3) 60⁰C for 30 sec
- 4) 68⁰C for 1 min
- 5) Repeat 2-4 for 25 cycles
- 6) 68⁰C for 5 min
- 7) 4⁰C forever
June 9th - Wet Lab
- Created cell culture with selection construct (contains ZFB, His3, pyrF on plasmid) and reporter RFP (this will be used to test positive control ZFs, cells fluoresce green when ZF binds)
- Picked colonies, grew in LB/amp liquid media until mid-log
- 3 mL of LB, 1.5 µL of 2000x amp
- Once mid-log reached, created glycerol stock, stored stock at -80⁰C.
300 µL bacteria, 1200 µL 80% glycerolThis should have been 1200 µL bacteria media, 300 µL 80% glycerol (Corrected 6/14/2011) (80% pure glycerol, 20% molecular grade water)
- Spiked new tubes of media with 25 µL bacteria from the mid-log tube to leave overnight
- Picked colonies, grew in LB/amp liquid media until mid-log
NOTE: reporter RFP did not grow to mid-log by end of day, will let grow overnight to saturation and continue creating glycerol stock tomorrow.
- Plated selection strain from gel stab onto tet plate.
- Began primer design for creating the kan/selection construct fusion.
June 9th - Bioinformatics
Today we focused on reacquainting and familiarizing ourselves with Python. We completed the parsing (reading in) of the sequence and amino acid data so that it is easy to work with: by substituting each amino acid abbreviation (ex. A, N) with its numeric equivalent (ex. 1, 14), we can use a lot of nice math comparisons instead of messy letter/"string" comparisons.
After that, we worked on counting the number of times each amino acid appears in each of the 7 positions (unfortunately given by -1,1,2,3,5,6,7), and counting the number of times amino acids are next to each other (ex. ACTQRNF has AC, CT, TQ, etc pairings). Taken overall, we found that L is overwhelmingly in position 5.
| Acid | -1 | 1 | 2 | 3 | 5 | 6 | 7 |
| A | 77 | 140 | 210 | 197 | 0 | 312 | 85 |
| C | 12 | 24 | 1 | 6 | 14 | 0 | 0 |
| D | 413 | 16 | 694 | 258 | 0 | 142 | 14 |
| E | 125 | 74 | 152 | 107 | 0 | 58 | 132 |
| F | 0 | 0 | 22 | 0 | 10 | 0 | 0 |
| G | 12 | 201 | 328 | 125 | 0 | 177 | 62 |
| H | 93 | 144 | 232 | 652 | 0 | 51 | 17 |
| I | 70 | 21 | 3 | 26 | 0 | 94 | 73 |
| K | 108 | 372 | 46 | 169 | 6 | 321 | 52 |
| L | 176 | 37 | 20 | 22 | 3325 | 75 | 55 |
| M | 36 | 54 | 5 | 28 | 0 | 31 | 10 |
| N | 23 | 150 | 129 | 940 | 0 | 182 | 61 |
| P | 3 | 298 | 77 | 7 | 0 | 36 | 8 |
| Q | 813 | 158 | 180 | 13 | 0 | 136 | 30 |
| R | 870 | 539 | 137 | 55 | 3 | 428 | 2517 |
| S | 99 | 970 | 859 | 278 | 0 | 140 | 12 |
| T | 243 | 134 | 223 | 350 | 0 | 834 | 83 |
| V | 166 | 26 | 27 | 115 | 0 | 341 | 146 |
| W | 19 | 0 | 13 | 0 | 0 | 0 | 0 |
| Y | 0 | 0 | 0 | 10 | 0 | 0 | 1 |
For pairings, we found patterns, but none as obvious as the L-in-position-5. Read this like a multiplication table: the intersection of L row and M column is how often that pairing was observed.
| ' | A | C | D | E | F | G | H | I | K | L | M | N | P | Q | R | S | T | V | W | Y |
| A | 10 | 0 | 99 | 55 | 0 | 29 | 122 | 20 | 32 | 332 | 2 | 59 | 55 | 63 | 255 | 87 | 24 | 43 | 0 | 0 |
| C | 0 | 0 | 15 | 0 | 0 | 3 | 0 | 0 | 0 | 5 | 0 | 0 | 6 | 0 | 31 | 6 | 14 | 0 | 0 | 0 |
| D | 99 | 15 | 94 | 92 | 0 | 39 | 62 | 6 | 84 | 342 | 15 | 120 | 55 | 42 | 277 | 290 | 87 | 21 | 0 | 8 |
| E | 55 | 0 | 92 | 42 | 0 | 34 | 77 | 1 | 38 | 141 | 2 | 39 | 4 | 29 | 134 | 28 | 90 | 26 | 0 | 1 |
| F | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 22 | 4 | 0 | 2 | 4 | 6 | 0 | 0 | 0 |
| G | 29 | 3 | 39 | 34 | 0 | 38 | 56 | 0 | 14 | 126 | 1 | 95 | 28 | 47 | 119 | 125 | 38 | 7 | 0 | 0 |
| H | 122 | 0 | 62 | 77 | 0 | 56 | 118 | 9 | 103 | 498 | 4 | 88 | 24 | 26 | 87 | 159 | 70 | 2 | 0 | 0 |
| I | 20 | 0 | 6 | 1 | 10 | 0 | 9 | 6 | 8 | 95 | 3 | 5 | 17 | 3 | 62 | 16 | 17 | 4 | 0 | 0 |
| K | 32 | 0 | 84 | 38 | 0 | 14 | 103 | 8 | 84 | 386 | 24 | 44 | 19 | 102 | 269 | 163 | 113 | 22 | 1 | 0 |
| L | 332 | 5 | 342 | 141 | 0 | 126 | 498 | 95 | 386 | 174 | 32 | 686 | 16 | 112 | 362 | 276 | 875 | 360 | 0 | 8 |
| M | 2 | 0 | 15 | 2 | 0 | 1 | 4 | 3 | 24 | 32 | 0 | 7 | 2 | 11 | 39 | 14 | 3 | 1 | 0 | 0 |
| N | 59 | 0 | 120 | 39 | 22 | 95 | 88 | 5 | 44 | 686 | 7 | 8 | 36 | 28 | 120 | 254 | 84 | 34 | 1 | 0 |
| P | 55 | 6 | 55 | 4 | 4 | 28 | 24 | 17 | 19 | 16 | 2 | 36 | 0 | 3 | 29 | 150 | 21 | 13 | 11 | 0 |
| Q | 63 | 0 | 42 | 29 | 0 | 47 | 26 | 3 | 102 | 112 | 11 | 28 | 3 | 100 | 261 | 314 | 125 | 19 | 0 | 0 |
| R | 255 | 31 | 277 | 134 | 2 | 119 | 87 | 62 | 269 | 362 | 39 | 120 | 29 | 261 | 618 | 343 | 504 | 281 | 0 | 0 |
| S | 87 | 6 | 290 | 28 | 4 | 125 | 159 | 16 | 163 | 276 | 14 | 254 | 150 | 314 | 343 | 592 | 173 | 91 | 0 | 0 |
| T | 24 | 14 | 87 | 90 | 6 | 38 | 70 | 17 | 113 | 875 | 3 | 84 | 21 | 125 | 504 | 173 | 154 | 28 | 0 | 0 |
| V | 43 | 0 | 21 | 26 | 0 | 7 | 2 | 4 | 22 | 360 | 1 | 34 | 13 | 19 | 281 | 91 | 28 | 12 | 0 | 0 |
| W | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Y | 0 | 0 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
June 10th - Wet Lab
- What we learned today: don't put E. coli plates in the -20C freezer!
- Observed a well populated selection strain plate and placed it in the 4C refrigerator
- Took the selection construct culture and extracted the plasmid through miniprep
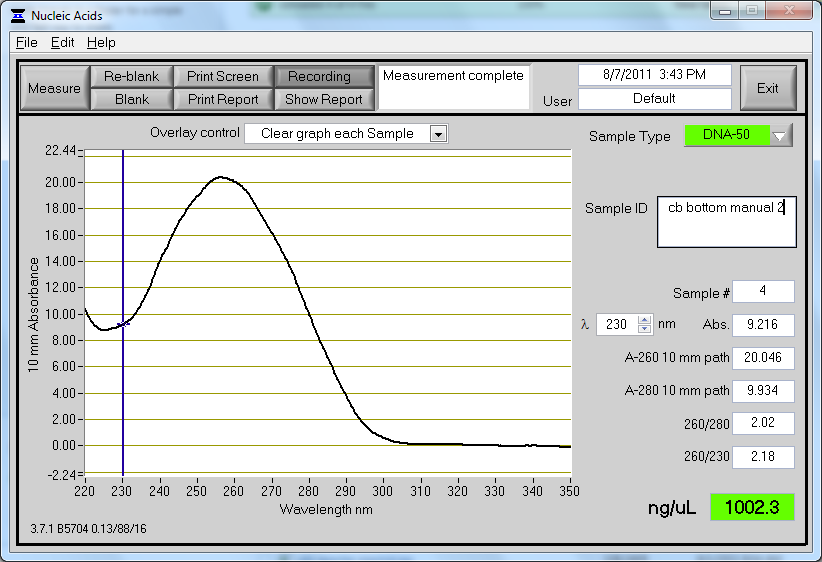
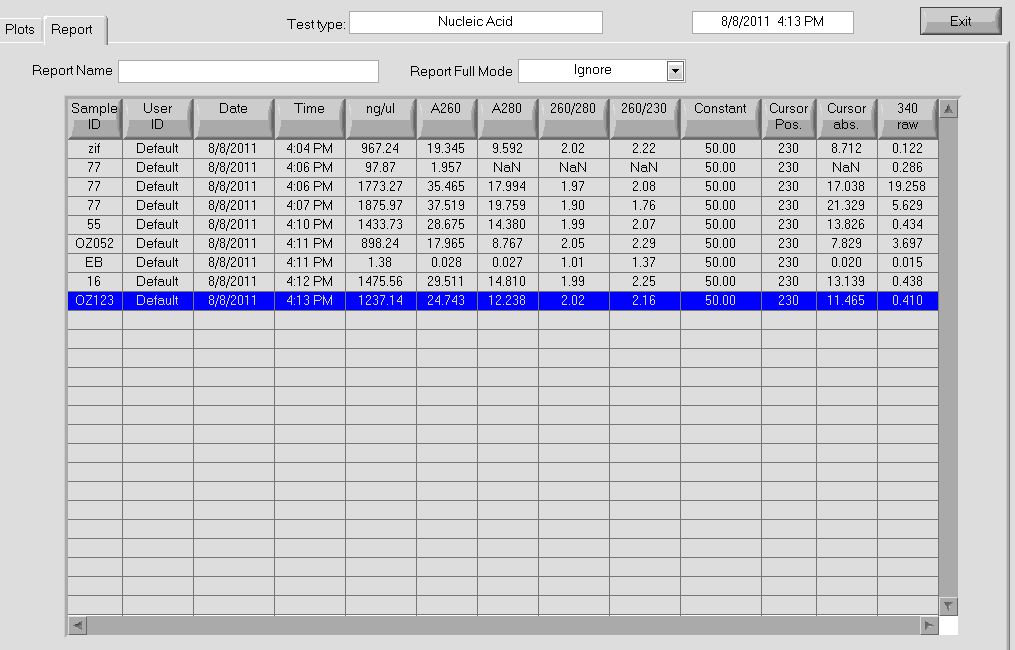
- Observed 260/280 ratio of 1.90 and 1.88 through Nanodrop
- Observed concentrations of 87.7 and 100.6 ng/µL through Nanodrop
- Made 10 new agar plates with LB and amp
June 10th - Bioinformatics
Visualizations
We spent the first few hours today making cool visualizations and graphs of the data we found on the 9th: heatmaps turned out to be an annoying limitation of Python, so a Python/R hybrid was used, and bar charts were made exclusively in Python. See the dropbox for our pretty (and hopefully informative compared to spreadsheets) charts/graphs.
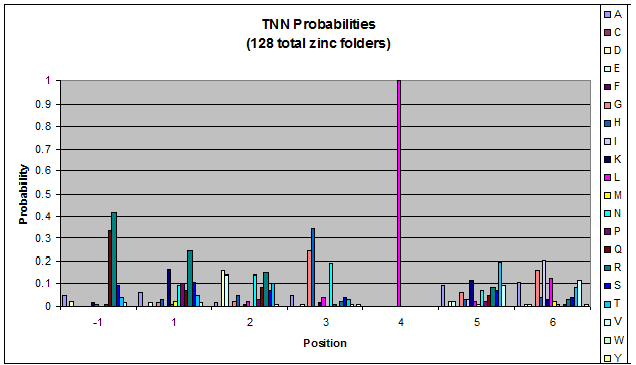
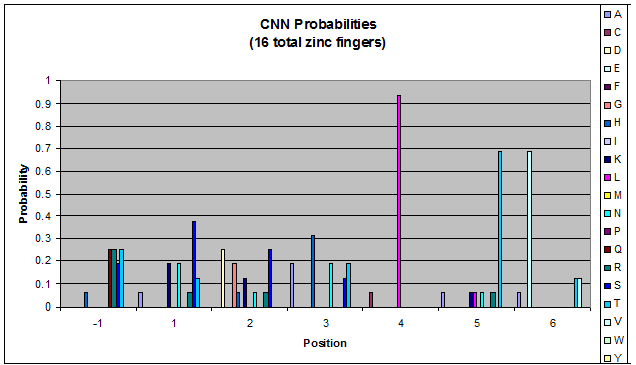
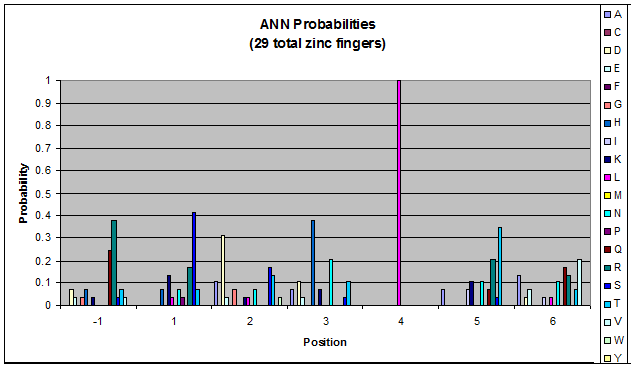
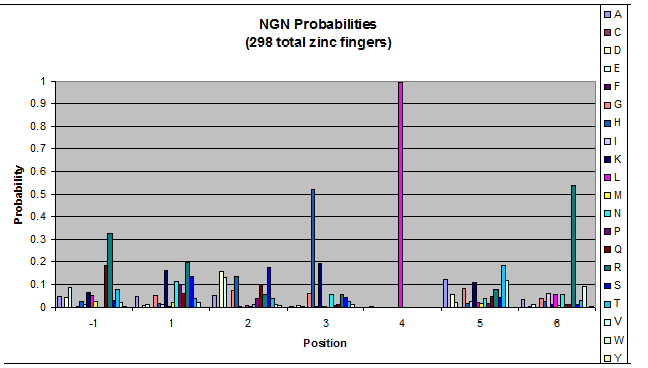
We then started work on TNN and GNN properties specifically (essentially repeating the June 9th work, but confined to smaller data sets). There are some differences between TNN and GNN: see graphs in dropbox. We decided that there was not enough data for fingers that bind to ANN and CNN triplets to perform significant analysis on it.
- Overall, similar color clusters are found in the heatmaps. In all cases, L and N are often placed consecutively on the helix. There are fewer clusters of high frequency when looking at TNN binders.
We then, using the theorized framework from a paper (2011 Persikov [http://iopscience.iop.org.ezp-prod1.hul.harvard.edu/1478-3975/8/3/035010/]), tried to match amino acid binding to each base pair to see if there was a pattern. See dropbox document .../bioinformatics/Binding Frequency for that data. There's a lot of it.
Properties of amino acids
We then worked on finding properties of the each position (hydrophobic/phillic, non/polar):
Hydrophilic vs Hydrophobic
| Position | Very Phobic | Hydrophobic | Neutral | Hydrophillic |
| 6 | 285 | 85 | 204 | 2782 |
| 5 | 542 | 312 | 1334 | 1169 |
| 4 | 3334 | 14 | 0 | 9 |
| 3 | 191 | 203 | 1417 | 1536 |
| 2 | 91 | 211 | 1819 | 1236 |
| 1 | 138 | 164 | 1604 | 1451 |
| -1 | 468 | 90 | 1257 | 1542 |
Polar vs Nonpolar
| Position | Polar | Nonpolar |
| 6 | 2917 | 440 |
| 5 | 2290 | 1067 |
| 4 | 9 | 3348 |
| 3 | 2830 | 527 |
| 2 | 2652 | 705 |
| 1 | 2555 | 802 |
| -1 | 2784 | 573 |
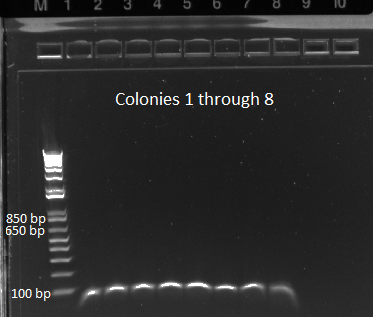
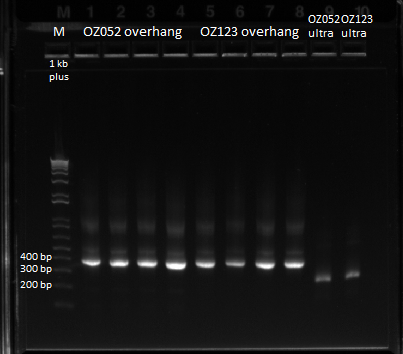
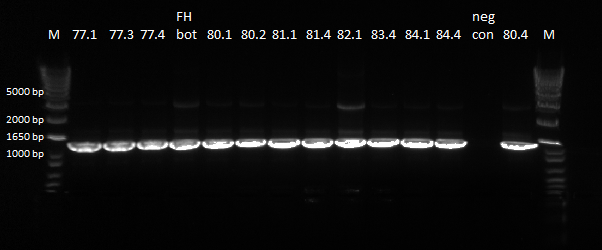
June 13th - Wet Lab
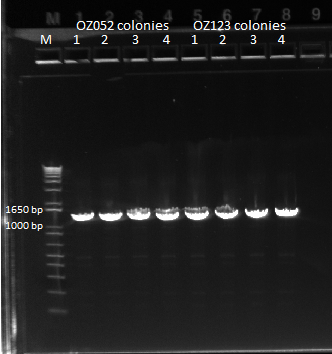
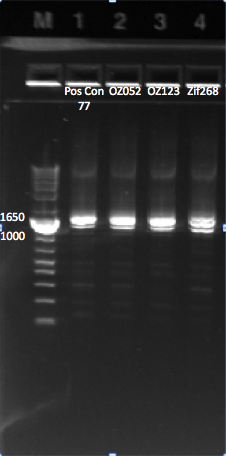
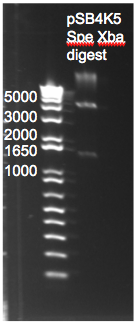
The control zinc fingers OZ052 and OZ123 were amplified with overhanging primers to allow its insertion into the Wolfe plasmid:
Overhang PCR for ultramers: the template was the product of the ultramer PCR (see 6/8/11), and several concentrations were used
In all the tubes:
- 5 µL Pfx amplification buffer
- 1.5 µL dNTPs
- 1 µL MgSO4
- 0.4 µL polymerase
- 38.1 µL ddH2O
- 1.5 µL OZ052_up and 1.5 µL OZ052_down OR 1.5 µL OZ123_up and 1.5 µL OZ123_down
In OZ052 (1) and OZ123 (1):
- 1 µL of ultramer PCR product
In OZ052 (1:10) and OZ123 (1:10):
- 1 µL of a 1 in 10 dilution of ultramer PCR product
In OZ052 (1:100) and OZ123 (1:100):
- 1 µL of a 1 in 100 dilution of ultramer PCR product
Parameters:
- 94⁰C for 5 min
- 94⁰C for 15 sec
- 55⁰C for 30 sec
- 68⁰C for 30 sec
- Repeat steps 2-4 for 25 cycles
- 68⁰C for 5 min
- 4⁰C forever
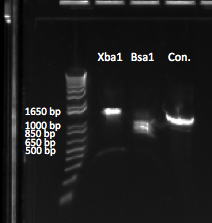
Gel to verify proper amplification (1% agarose gel, 10 µL 1 kb ladder, 120 V):
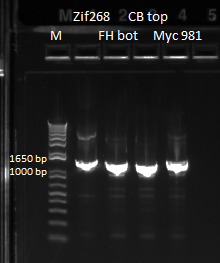
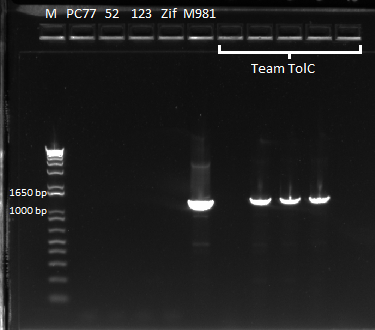
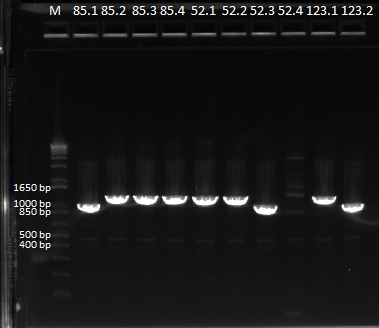
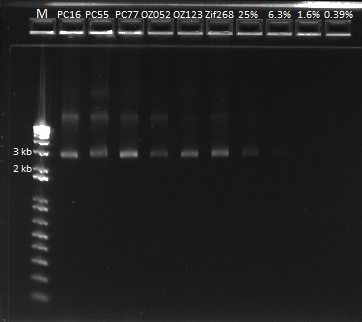
The OZ052 lanes (1-3) had bands at the proper length (328 bp) at all three concentrations, although there were several fainter bands likely from side products. Only the undiluted OZ123 lane showed any bands, and from the faint band at 328 and the stronger band around 250 it appears that the PCR did not work well, and the majority of the product was the ultramer from the first PCR.
PCR around vector: the template used was the Wolfe selection construct plasmid miniprepped 6/10/11 (100.6 ng/µL stock)
Reagents the same as above except:
- 1.5 µL of Wolfe_F and 1.5 µL of Wolfe_R primers to each tube
- plasmid tube (1 ng) given 1 ng of template (1 µL of a 1 in 100 dilution)
- plasmid tube (10 ng) given 10 ng of template (1 µL of a 1 in 10 dilution)
Parameters same as above except:
- elongation (step 4) 5 minutes (vector approximately 5 kb)
Gel to verify proper amplification (1% agarose, 10 µL 1 kb ladder, 170V)
There were no bands of the correct size in the lanes. The only band that appeared was a faint, short band in one lane that likely was a primer. Since the DNA ladder worked, the problem likely was not with the electrophoresis but with the PCR reaction, perhaps due to issues with the primers.
Gel images
June 13th - Bioinformatics
Today we started work on a program to statistically generate possible sequences.
The four functions needed to do this are:
- generate(matrix, pseudocounts (lambda), dependency tuples)
- takes a matrix of zinc-finger AA position counts, a list of dependent amino acid pairs, and a pseudocount multiplier and generates a list of potential amino acid sequences weighted by independent and dependent probabilities
- add_pseudo(dependent matrix row,independent matrix row)
- given a matrix row of dependent counts (i.e. how many times 'a' occurs at position n when 'b' is set to some AA at position m) and a row of independent matrix counts (how many times 'a' occurs at n regardless of b's AA) return an adjusted matrix row, based on the dependent matrix row, that has pseudocounts added to the values that are empty in the dependent matrix row but filled in the independent matrix row.
- generate_indep(matrix)
- randomly pick an amino acid, given a matrix row, from a weighted random distribution based on the values in the row
- generate_dep(indep_row, dep_row, lambda)
- add pseudo counts (call add_pseudo) and generate a dependent random call for a position (using generate_indep on the adjusted matrix)
June 14
Wet Lab
- Made four LB-based media solutions, and later created glycerol stocks from these and placed in -80⁰C freezer
- Selection strain (ΔHis3ΔPyrFΔrpoZ) in 3 mL LB and 3µL of 1000x Tet solution stock
- Selection strain (ΔHis3ΔPyrFΔrpoZ) in 3 mL of LB only solution stock (control)
- Kan cassette (pZE22G) in 3 mL of LB solution stock and 3 µL of kanamycin solution stock
- Lambda Red (pKD42) in 3 mL of LB and 3 µL Amp solution stock
- For all of these stocks, we tried to grow all to mid-log and then place them in 1,200 µL of culture and 300 µL of 80 % glycerol solution (This is the correct protocol for creating a glycerol stock; refer to June 9th)
- We were only able to get the kan cassette to mid-log and created glycerol stock of the kan cassette
- Observations included contamination of a pKD46 liquid culture, and we are leaving Lambda Red and both solutions with the selection strain for overnight growth
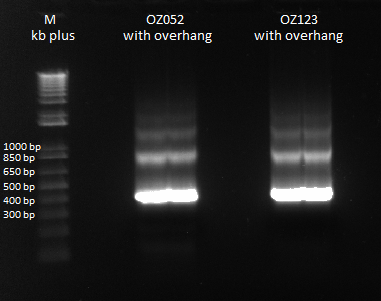
- Ran 1% gel (150V) with the rest of the OZ123 and OZ052 overhang PCR samples
- Used better ladder today, less the 1 kb ladder
- Bands followed the same pattern as the gel run on 6/13/11
- Used gel extraction to obtain the correct OZ123 and OZ052 PCR product from the gel
- Used Qiagen quick gel extraction kit
- OZ052 (from undiluted lane): 7.0 ng/µL, 260/280=2.42 (Note: this sample had a strange yellow substance in the column, may have been contaminated)
- OZ052 (from undiluted lane): 12.0 ng/µL, 260/280=2.02
- OZ052 (from 1:10 dilution): 10.8 ng/µL, 260/280=2.04
- OZ052 (from 1:10 dilution): 15.2 ng/µL, 260/280=1.82
- OZ052 (from 1:100 dilution): 20.6 ng/µL, 260/280=2.17
- OZ123 (from undiluted lane): 6.3 ng/µL, 260/280=2.17
- PCR the backbone fragment of the plasmid using Wolfe_R and Wolfe_L primers and a lower annealing temperature than before due to the lower melting point of Wolfe_L
- Reagents:
- 22µL Invitrogen Platinum PCR supermix
- 1µL template from a 1 in 100 dilution (1 ng)
- 1µL Wolfe_F and 1µL Wolfe_R
- 94°C for 30 s
- 94°C for 30 s
- 53°C for 30 s
- 70°C for 5 min
- Previous three steps repeat 30 times
- 70°C for 5 min
- 4°C forever
- Reagents:
- Performed a restriction enzyme digestion on the selection construct plasmid using EcoRI to test for presence/absence of inserted selection construct
- 1µL EcoR1
- 1µL buffer 4
- 2µL backbone plasmid
- 6µL ddH2O
- Incubate at 37 degrees for 90 min
- There is only one EcoR1 site (GAATTC) in the plasmid, so we should see 1 band at about 5kb
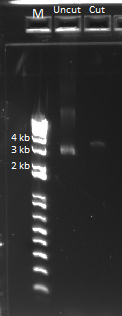
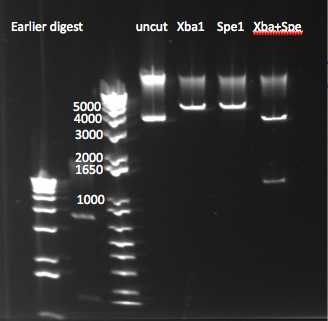
- Ran a gel (1%, 170 V) on the backbone fragment (Wolfe primers) PCR product and restriction digestion result
- Observations: the EcoR1 digest produced the expected band of around 5kb. The backbone did produce a 5kb band but also had a secondary smaller product, perhaps due to one of the primers annealing to a sequence that is a close match to its target.
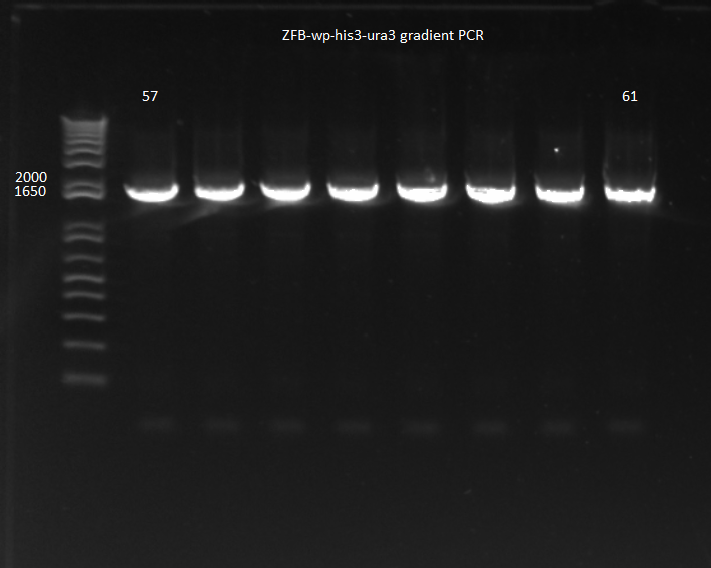
- Began gradient PCR on the selection strain backbone with Wolfe_R and Wolfe_F primers because of the large difference in melting temperatures between the two
- Set the annealing temperatures within the PCR to go from 50-57 C and ran with 5 minute extension phases at 70 C
- Ran overnight
Today's Gel Images
June 14 - Bioinformatics
We finished writing the generate function, and now have a working sequence generator. We also began more in-depth research into the 2011 work by Persikov which deals with how the zinc finger binds to DNA. He predicts several relations which we should be able to test.
Persikov sent us his SVM code (used to calculate the probability of a sequence binding to given DNA), so we also worked on adapting this to use when narrowing our sequences to those most likely to work.
- There are four canonical amino acid-base interactions involved in zinc finger interactions.
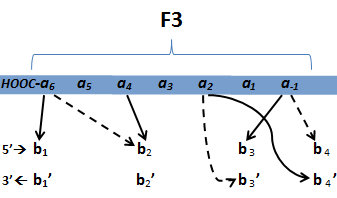
- Amino acids in positions -1, 3, and 6 on the helix are known to interact with the 3 bases of the triplet. In addition, the amino acid in position 2 interacts with the upstream base of the complementary strand (Klug 2010).
- In addition, Persikov (2011) has proposed novel interactions between these four amino acids based on his analysis of zinc finger binding data up to 2005.
- Persikov uses the information between these four amino acid-base interactions in his SVM, to determine whether a finger would be a good binder to a particular DNA sequence.
- In order to use his program, we need to convert the helices that are created by the generator into a format the SVM accepts.
- The SVM only considers the four canonical interactions. It assigns a numerical value to each possible amino acid-base combination. The program that converts our data into a format the SVM accepts creates a string with these numerical values based on Persikov's key. (See this page for more details on this program.)
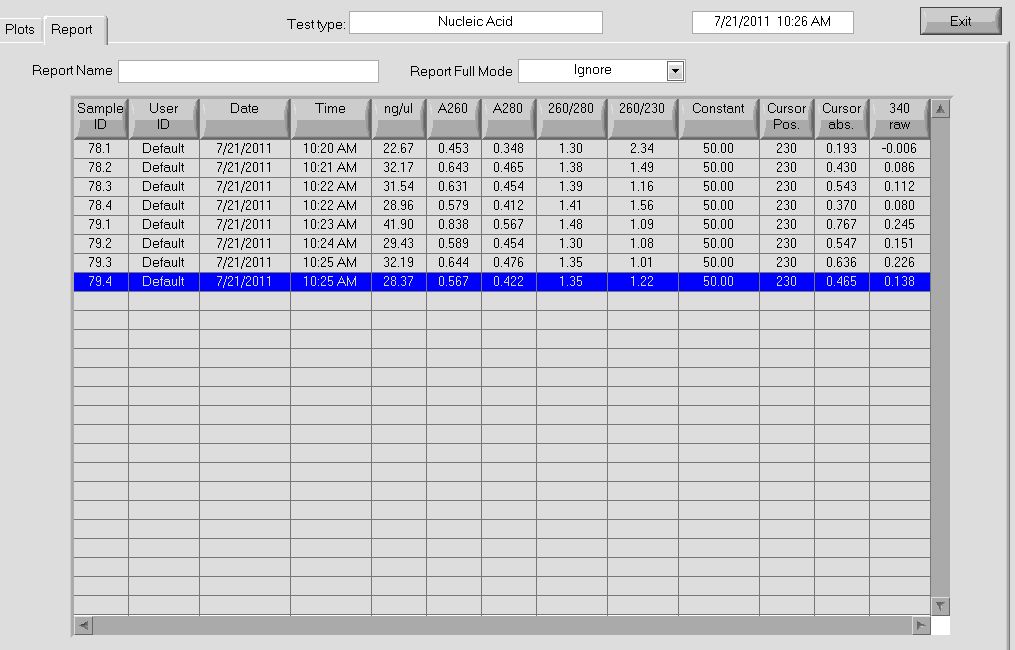
June 15th - Wet Lab
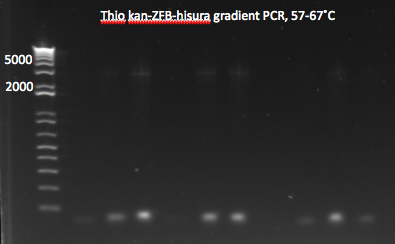
Gradient PCR: 5µL of each PCR product were run on a 1% gel. No bands appeared: the PCR appears to not have worked.
Selection construct: bacteria containing the selection construct (plasmid containing ZF, omega subunit, ZFB, His3, URA3, etc.) was made into a glycerol stock (see 6/14 and 6/9), miniprepped, and used for PCR:
- Miniprep: used Qiagen kit
- 82.5ng/µL, 260/280=1.99
- 91.1ng/µL, 260/280=2.01
- 77.9ng/µL, 260/280=1.98
- PCR:
- PCR used to amplify section of plasmid containing zinc finger binding site, weak promoter, His3, and URA3 (with homology to join it to kan cassette)
- Reagents
- zinc finger binding site and weak promoter, selection construct plasmid:
- 1µL ZFB-wp-f (5µM) (made by 1:20 dilution of 100µM stock)
- 1µL ZFB-wp-hisura-r (5µM) (made by 1:20 dilution of 100µM stock)
- 1µL selection construct (1:100 dilution of overnight culture)
- 22.5µL of invitrogen's Platinum PCR SuperMix
- PCR used to amplify the kan cassette
- Reagents
- KAN cassette, pZE22g plasmid:
- 1µL hisura-kan-f (5µM) (made by 1:20 dilution of 100µM stock)
- 1µL kan-r (5µM) (made by 1:20 dilution of 100µM stock)
- 1µL pZE22g (1:100 dilution of glycerol stock culture)
- 22.5µL of invitrogen's Platinum PCR SuperMix
- Parameters:
- 1) 94°C for 2 min (denature template, activate enzyme)
- 2) 94°C for 30 sec (denature)
- 3) 53°C for 30 sec (anneal)
- 4) 72°C for 2 min (extend)
- 5) Repeat 2-4 for 25 cycles total
- 6) 72⁰C for 5 min
- 7) 4°C forever
PCR Purification:
- Used the Qiagen PCR purification kit and instructions in order to purify the Kan cassette and selections construct PCR products
- Nanodrop the purification results and observed 3 ng/µL for Kan cassette and 29.8 ng/µL for ZFB-wp-His3: purification did not work well, especially for the kan
Selection strain (ΔHis3ΔPyrFΔrpoZ):
- saturated overnight culture was inoculated again: 3mL LB, 3µL tet, 30µL of overnight culture, at 37C until mid-log
- glycerol stock
- For transformation tomorrow we grew up pKD42 in 3 mL of LB, 1.5 µL of ampicillin(2000x) and one colony at 30 C
- Also grew up more of the selection strain so it will be ready for electroporation transformation
Gel
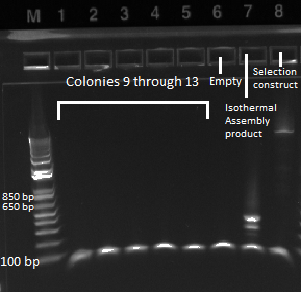
- Ran gel with Kan cassette and selection construct (Binding site, His3, and URA3)
- Observations successful and image below
- Used 1 kb plus ladder
PCR Overlap
- since the purification was not very successful, we used 3µL saved from the original PCR product
- Procedure
- 25µL of 2x Phusion Master Mix
- 1 µL of ZFB-wp-HisURA-R (100µM)
- 1µL of HisURA-Kan-F (100µM)
- 21 µL of water
- 1 µL Kan template and 1 µL of ZFB-wp-His3-URA3
- 4 tubes
- Both undiluted
- Both 1:10 dilution
- Both 1:100 dilution
- Both 1:1000 dilution
- 4 tubes
- Protocol
- 98 C for 30 s
- 98 C for 10 s
- 53 C for 30 s
- 72 C for 3 min
- Repeat steps 2-4 for 24 more cycles
- 72 C for 5 min
- 4 C 4EVA!!!
June 15th - Bioinformatics
- We continued research into Persikov's and others' work on binding.
- We worked on using the OPEN data to test Persikov's binding-predicting program: SVM
- Justin continued work on a sequence-finding program, the most up to date version can be found in the Dropbox under code/zfsitefinder.html.
- Justin and Will found 10 candidate sequences across 4 diseases that hopefully should encompass a good amount of diversity in terms of expanding the ZF library. These sequences can be found in the table below, with more details here.
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,403,001-153,407,000 | 370 | GTATTTGTT | GGGCCTGCT | N/A | N/A |
| Colorblindness | chrX:153,403,001-153,407,000 | 3627 | GCTGGCTGG | GCGGTAATG | EGSGLKR.EAHHLSR.####### | RRDDLTR.QRSSLVR.####### |
| Cystic Fibrosis | chr7:117,074,084-117,089,556 | 14767 | GCAGGTGAT | AAAGAGCCC | QNGTLGR.EAHHLSR.####### | N/A |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 14001 | GGCTGAGAC | GGAGTCCTG | ESGHLKR.QREHLTT.####### | QTTHLSR.DHSSLKR.####### |
| Tay-Sachs | chr15:72,674,944-72,688,031 | 5888 | GTCTGGTCA | TCAAACTCC | DRSSLRR.RREHLTI.####### | N/A |
| Pancreatic Cancer | chr7:117,074,084-117,089,556 | 1739 | GATCAAGCT | GTTTCAGTG | N/A | N/A |
- We collected 15 alternative zinc finger backbones (different from zif268 backbone) and their corresponding base sequences. Many of these were from Persikov 2011 and all binding sequences were confirmed on the [http://www.pdb.org/pdb/home/home.do Protein Data Bank website]. The zinc finger PDB ID's and related links are:
| PDB ID | Binding Sequence | Link |
|---|---|---|
| 1F2I | ATGGGCGCGCCCAT | [http://www.pdb.org/pdb/explore/explore.do?structureId=1F2I] |
| 1G2D | GACGCTATAAAAGGAG | [http://www.pdb.org/pdb/explore/explore.do?structureId=1G2D] |
| 1G2F | TCCTTTTATAGCGTCC | [http://www.pdb.org/pdb/explore/explore.do?structureId=1G2F] |
| 1MEY | ATGAGGCAGAACT | [http://www.pdb.org/pdb/explore/explore.do?structureId=1MEY] |
| 1TF6 | ACGGGCCTGGTTAGTACCTGGATGGGAGACC | [http://www.pdb.org/pdb/explore/explore.do?structureId=1TF6] |
| 1UBD | AGGGTCTCCATTTTGAAGCG | [http://www.pdb.org/pdb/explore/explore.do?structureId=1UBD] |
| 1TF6 | ACGGGCCTGGTTAGTACCTGGATGGGAGACC | [http://www.pdb.org/pdb/explore/explore.do?structureId=1TF6] |
| 1YUI | GCCGAGAGTAC | [http://www.pdb.org/pdb/explore/explore.do?structureId=1YUI] |
| 2DRP | CTAATAAGGATAACGTCCG | [http://www.pdb.org/pdb/explore/explore.do?structureId=2DRP] |
| 2GLI | TTTCGTCTTGGGTGGTCCACG | [http://www.pdb.org/pdb/explore/explore.do?structureId=2GLI] |
| 2I13 | CAGATGTAGGGAAAAGCCCGGG | [http://www.pdb.org/pdb/explore/explore.do?structureId=2I13] |
| 2KMK | CATAAATCACTGCCTA | [http://www.pdb.org/pdb/explore/explore.do?structureId=2KMK] |
| 2PRT | CGCGGGGGCGTCTG | [http://www.pdb.org/pdb/explore/explore.do?structureId=2PRT] |
| 2WBS | GAGGCGC | [http://www.pdb.org/pdb/explore/explore.do?structureId=2WBS] |
| 2WBU | GAGGCGTGGC | [http://www.pdb.org/pdb/explore/explore.do?structureId=2WBU] |
June 16th - Wet Lab
- There was totally a crazy bee hive outside today!!
Glycerol Stock pKD42
- Grew up pKD42 in 30 C and once reached mid-log created glycerol stock and placed in -80 refrigerator
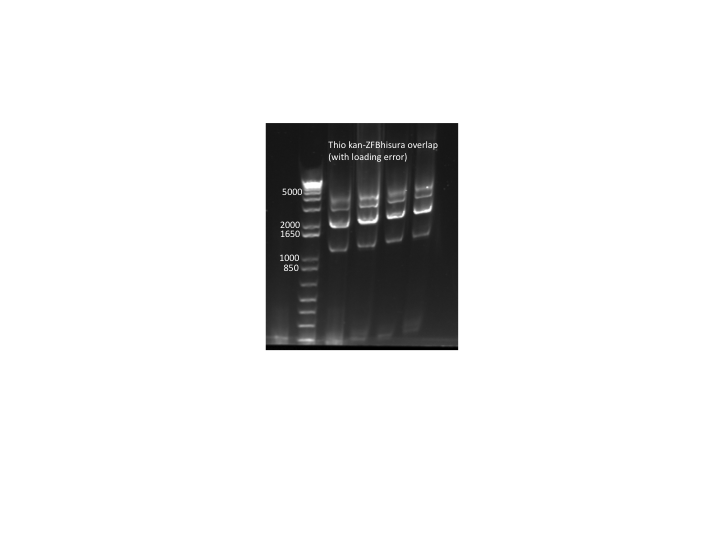
Overlap PCR gel
- Ran gel to test if overlap PCR that ran through the night worked, and it did not
- Used PCR product without purification which gives good explanation for why it didn't work
- PCR: Since the PCR done the previous day (6/15) we made a back up PCR using phusion mastermix (Finnzyme)
- PCR used to amplify section of plasmid containing zinc finger binding site, weak promoter, His3, and URA3 (with homology to join it to kan cassette)
- Reagents
- zinc finger binding site and weak promoter, selection construct plasmid:
- 1µL ZFB-wp-f (100µM) (taken directly from the primer tube)
- 1µL ZFB-wp-hisura-r (100µM) (taken directly from the primer tube)
- 2µL selection construct (1:100 dilution of overnight culture)
- 25µL Phusion High-Fidelity PCR Master Mix
- 21µL distilled water (for total volume of 50µL)
- PCR used to amplify the kan cassette
- Reagents
- KAN cassette, pZE22g plasmid:
- 1µL hisura-kan-f (100µM) (taken directly from the primer tube)
- 1µL kan-r (100µM) (taken directly from the primer tube)
- 2µL pZE22g (1:100 dilution of glycerol stock culture)
- 25µL of Phusion High-Fidelity PCR Master Mix
- 21µL distilled water (for total volume of 50µL)
- Parameters:
- 1) 94°C for 2 min (denature template, activate enzyme)
- 2) 94°C for 30 sec (denature)
- 3) 53°C for 30 sec (anneal)
- 4) 72°C for 2 min (extend)
- 5) Repeat 2-4 for 25 cycles total
- 6) 72⁰C for 5 min
- 7) 4°C forever
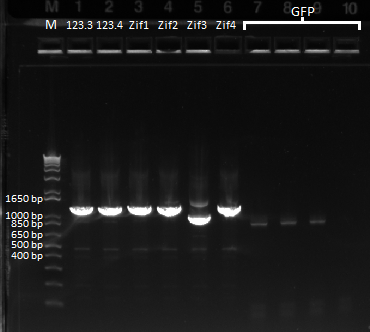
- Ran PCR product on a gel: bands of the correct size were observed, though the kan band was much fainter than the ZFB-wp-his3
- Repeat PCR (to get a higher concentration of the Kan cassette and ZFB-wp-his3 constructs)
- same as the above backup PCR (since it was successful), but to a 4x total volume of 200µL, compared to 50µL
- PCR products were run on a gel: the correct bands were observed--see image below ("second gel")
- PCR product purification: followed Qiagen kit instructions. Strangely, the conc. was 64.6 ng/µL for Kan, purity 2.09 (260/280) and for ZFB-wp-his3, the conc. was 23.7ng/µL and the purity was 1.92 (260/280).
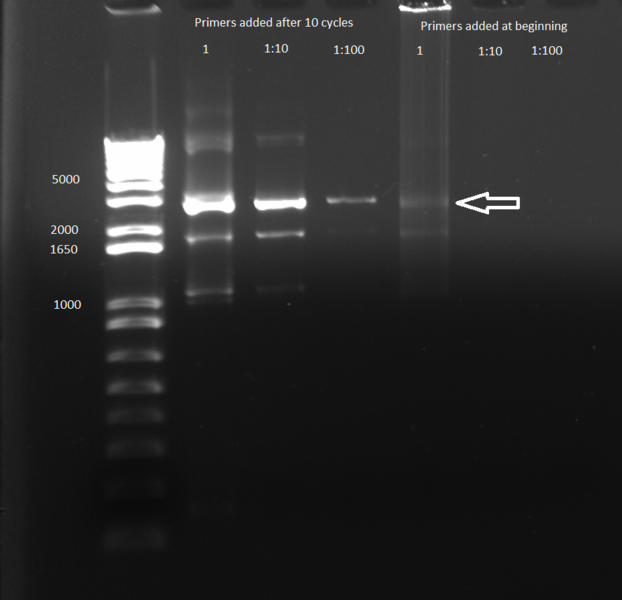
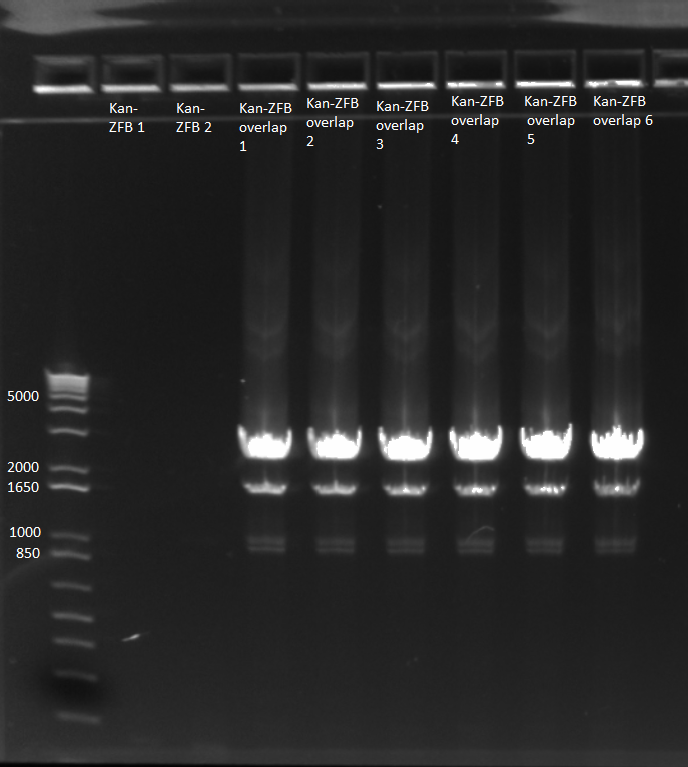
Overlap PCR: used the kan cassette (64.6 ng/µL) and ZFB-wp-his3-ura3 (23.7 ng/µL) purified above
- 1 µL of kan and 1 µL of ZFB-wp-his3-ura in each tube according to the following conditions:
- two tubes: undiluted
- two tubes: both diluted 1 in 10
- two tubes: both diluted 1 in 100
- 12.5µL Phusion master mix
- 8 µL ddH2O
- primers: 1.25 µL ZFB-wp-hisura_r (10 µM) and 1.25 µL hisura-kan_f (10 µM)
- we tried two different reaction types: one added the primers as usual before starting the PCR reaction, the other added the primers after 10 PCR cycles (allowing the polymerase to first use the overlapping kan and ZFB to elongate, and then the primers)
- Parameters for PCR starting with primers:
- (PCR machine 5, program name EXT3KB in IGEM folder)
- 1) 98°C for 1 min (denature template, activate enzyme)
- 2) 98°C for 15 sec (denature)
- 3) 65°C for 15 sec (anneal)
- 4) 72°C for 2 min (extend)
- 5) Repeat 2-4 for 30 cycles total
- 6) 72⁰C for 5 min
- 7) 4°C forever
- Parameters for PCR starting without primers:
- 1) 98C for 1 min
- 2) 98C for 15 sec
- 3) 65C for 15 sec
- 4) 72C for 1 min
- 5) back to step 2 for 10 cycles (PCR paused after 10 and primers added)
- 6) 98C for 15 sec
- 7) 65C for 15 sec
- 8) 72C for 2 min
- 9) back to step 6 for 20 cycles
- 10) 72C for 5 min
- 11) 4C forever
Transformation
- Used the selection strain (ΔHis3ΔPyrFΔrpoZ) cells at mid-log and attempted to place lambda red (pKD42) plasmid into the cell
- Procedure
- Keep on ice through out whole procedure before use of the electroporation machine
- Spin 1.5 mL of mid-log cells at 4 C for 1 minute at 18000 rcf (we created two tubes through the following steps)
- Discard supernatant and resuspend with 1 mL of cold water
- Spin again and repeat for a second water wash
- With each wash, try to get as much supernatant out as possible(even use pipette) because don't want salts to interfere with the electrical pulse
- Resuspend pellet with 50 µL of cold water
- Add 1 ng of pKD42 to one of the tubes and 45 ng of pKD 42 to the other
- Take all of the liquid in each tube and place in two separate cuvettes for electroporation
- Make sure the electroporation machine is on the right setting (for the cuvettes we used today it was "Ec2")
- Wipe off all water on the side of the cuvette
- Have 1 mL of LB in hand and after pulsing, immediately put LB in cuvette
- Transfer to culture tube and place in 30 C for 2 hours
- Make 4 LB/amp plates and spread E. coli using glass beads:
- Plate 1: 10 µL of 1 ng culture
- Plate 2: take 700 µL of 1 ng culture, spin down and remove supernatant, resuspend in about 30 µL of LB and plate
- Plate 3: 10 µL of 45 ng culture
- Plate 4: take 700 µL of 45 ng culture, spin down and remove supernatant, resuspend in about 30 µL of LB and plate
- Grow overnight at 30 C
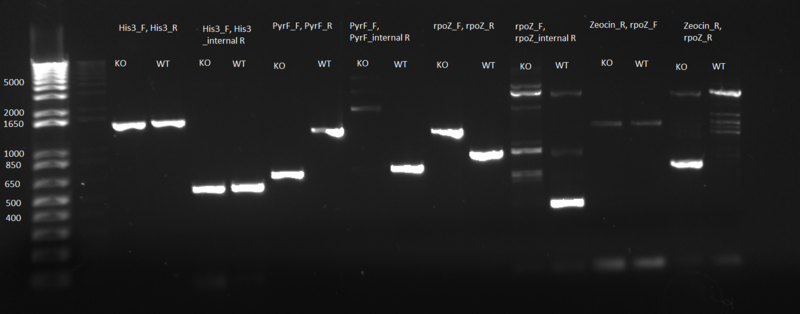
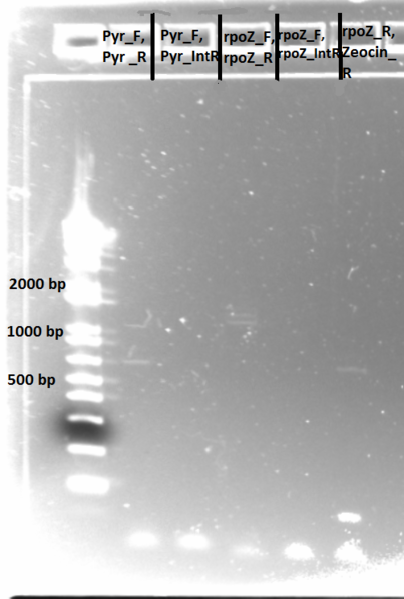
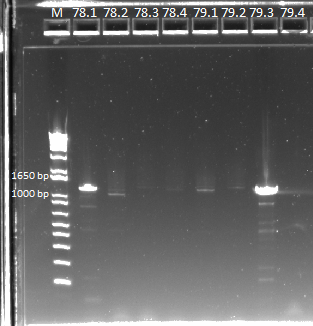
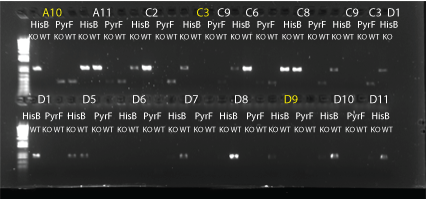
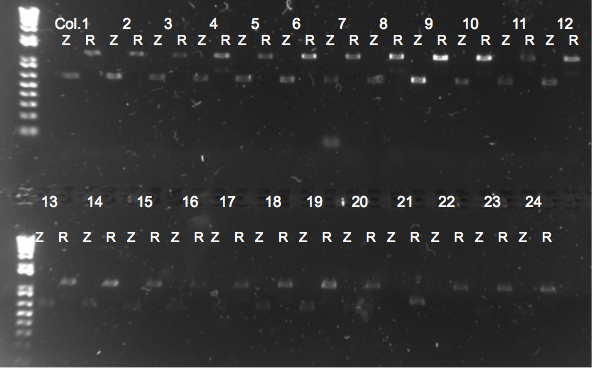
PCR to confirm knockouts of selection strain
- this PCR was to confirm that the ΔHis3ΔPyrFΔrpoZ was indeed a knockout for the His3, PyrF, and rpoZ genes
- each primer set was used for two conditions: wild-type (we used a pKD42 culture) and knockout (ΔHis3ΔPyrFΔrpoZ culture, left over from transformation)
- 1 µL of either wt or ko template, diluted 1:20
- 12.5 µL Phusion master mix
- 1.25 µL of each 10µM primer:
- test for His3:
- 1)His3_F, His3_R
- 2)His3_F, His3_internalR
- test for PyrF:
- 3)PyrF_F, PyrF_R
- 4)PyrF_F, PyrF_internalR
- test for rpoZ:
- 5)rpoZ_F, rpoZ_R
- 6)rpoZ_F, rpoZ_internalR
- test for Zeocin (there are two primer sets because we don't know what orientation the Zeocin gene is in)
- 7)Zeocin_R, rpoZ_F
- 8)Zeocin_R, rpoZ_R
- test for His3:
- ddH2O up to 25 µL
- Parameters:
- 98 C for 5 min
- 98 C for 10 sec
- 65 C for 25 sec
- 72 C for 45 sec
- cycle 30 times
- 72 C for 5 min
- 4 C forever
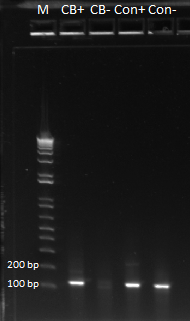
- Results: ran PCR products out on 1% gel (see below). There were some nonspecific bands, but the PyrF and rpoZ genes do appear to be knocked out in the selection strain. His3, however, looks like it's still present--we'll test again to confirm.
June 16 - Bioinformatics
- Research Targets
- Clinically relevant targets
- Existing ZFs that bind under-represented triplets
Updating our programs
- Many of our current programs currently look at overall data or data based on specific DNA triplets (for example: 'GAT' or 'AAA'). However, in order to more easily understand some of the patterns that occur in the datasets, we want to examine broader subsets of data. For example, do different patterns appear when looking at fingers that bind to 'GNN' triplets versus 'NGN' triplets (where 'N' represents any of the 4 bases)?
- We added the capability for our programs to accept inputs with the variable 'N' by using regular expressions.
- We can now create lists of the zinc fingers that bind to any triplet, and create interaction matrices and frequency tables for any triplet input.
- We added the capability for our programs to accept inputs with the variable 'N' by using regular expressions.
June 17th - Wet Lab
Update on selection strain knockout status: We are trying to reach Addgene to check how His3 was knocked out---instead of deleting the gene, they may have simply introduced an early stop codon. If that's the case, our gel would have the correct bands because the primers we designed can only show whether a deletion or insertion was in that locus.
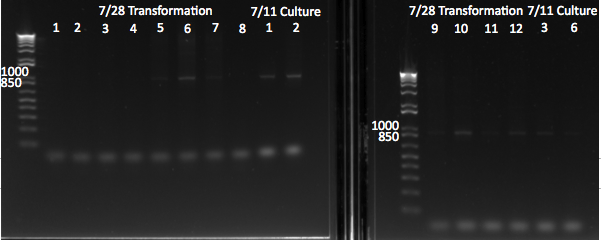
Transformation results: successful!!
- The only plate with colonies was the one plated with 700 µL of cells transformed with 45 ng of pKD42
- Chose a colony to grow in 3mL LB, 1.5µL amp, 30C; make glycerol stock with mid-log cells
- Plate with colonies at 4C
Miniprep of pZE22G: (to have the plasmid containing the kan cassette on hand)
- used 2 tubes of 1.5mL overnight culture, followed Qiagen kit instructions
- 38.0 ng/µL, 260/280=1.99
- 27.8ng/µL, 260/280=2.02
Overlap PCR gel and extraction: 1%, 150V
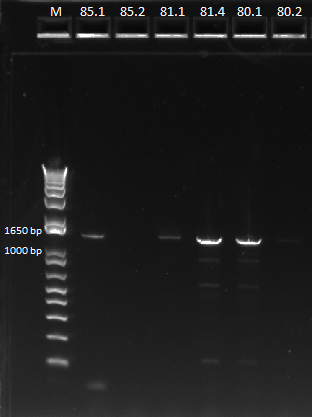
- Results: adding the primers in after 10 cycles was much more successful than adding them at the beginning, and all three dilutions showed the expected product band (about 2.5kb). The rest of the 1:10 dilution will be run on a gel and extracted.
- 11.2ng/µL, 260/280=2.10
June 17 - Bioinformatics
Goals
- Make BB Database in program-readable format ✓
- Edit out BB with incomplete helices ✓
- GNN, TNN, CNN, ANN frequencies
- Targets (5-10; 8) x Backbones (???) x Helices (≥500)=55,000
- Backbones: similar, but not too similar to zif268; more than 1-2 aa changes, but <10
- Helices fixed based on our program-- eventually saturates and levels out
- Graph: # of var (# of tries by the computer) vs. % space covered
Options for Target DNA Sequences / ZF Helices
- F3(known) / F2(known) / F1(novel)
- F3(known) / F2(SNP in b1 position) / F1(known)
- F3(unknown) / F2(unknown) / F2(unknown)
- Excluded Rare Codons (for E. coli)CodonUsage OpenWetWareCodonUsage NIHRareCodonCalculator:
- CTA
- ATA
- CCC
- CGA
- CGG
- AGA
- AGG
- GGA
- GGG
References
- Persikov2011 pmid=21572177
- CodonUsage http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/in-vitro-genetics/codon-usage.html
- OpenWetWareCodonUsage http://openwetware.org/wiki/Escherichia_coli/Codon_usage
- NIHRareCodonCalculator http://nihserver.mbi.ucla.edu/RACC/
June 20th - Wet Lab
- Grew up colony of the selection strain with pKD46 in an attempt to reach mid-log and create glycerol stock
- Unable to reach mid-log, so going to leave growing over night and use saturated culture tomorrow
- Determined primers in order to piece together the omega subunit and ZFP genes into the pZE21G plasmid (spec cassette)
- Ran PCR on His3 locus and sent to GENEWIZ to be sequenced
- used the same procedure as the earler WT/KO PCR, but with 1µL undiluted template and only His_F and His_R primers
- ran 3 reactions and sent in three primers (His_F, His_R, His_internalR)
June 20th - Bioinformatics
Goals for the week
- Finish designing the chip, by Wednesday hopefully
- Need chip order out, takes 4 weeks
- Need all sequences by Friday!!!
- FIRST PRIORITY: If we can get Persikov to work, good!
- Step one: get results he’s published, get the web app to "work" with his data, then OPEN data, and finally our data
- Brainstorming session (tomorrow?) to decide how many targets/sequences
- Determine the importance of the first/second/third nucleotide positions
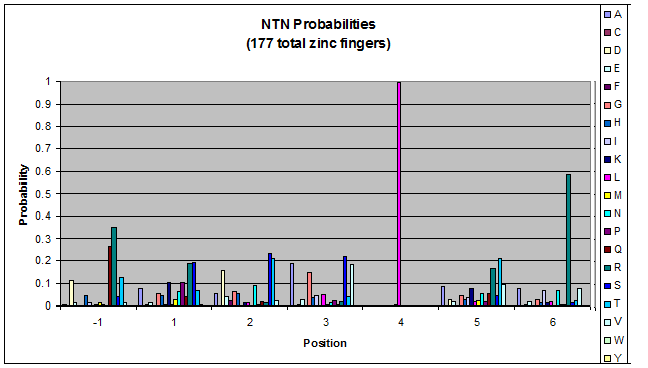
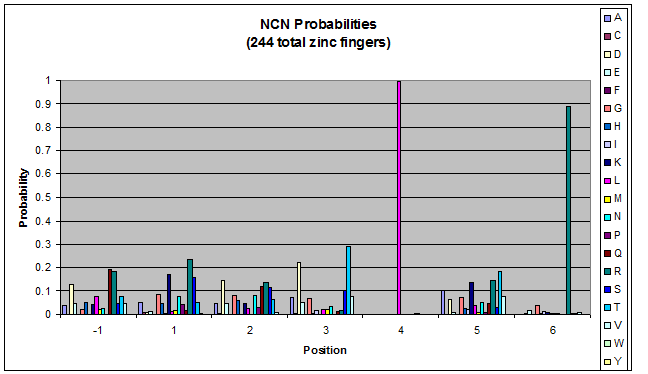
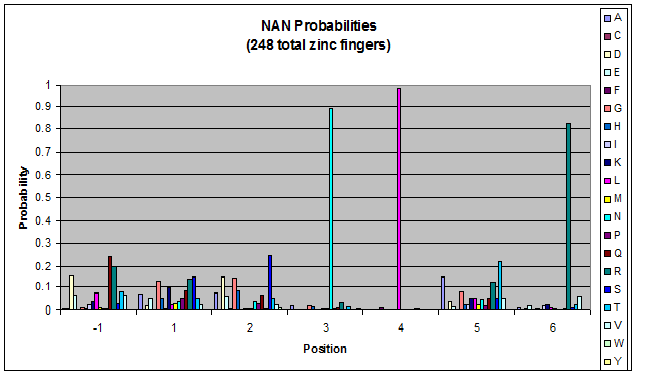
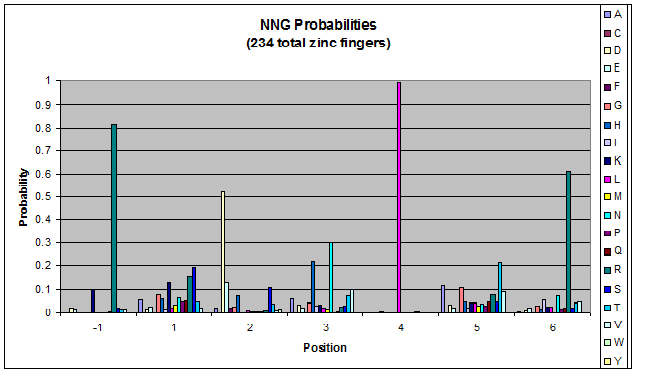
- Look at NGN, NTN, NAN, NCN (Not just GNN, etc.)
- Pick a particular GNN, plot vs. TNN- is there a pronounced difference in position 1, or -1?
Today
- Testing Persikov's Data for validation
- Persikov v. himself ✓
- Persikov v. OPEN
- Persikov v. our sequences
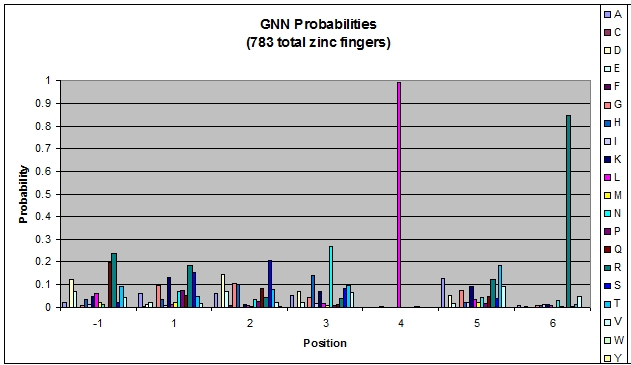
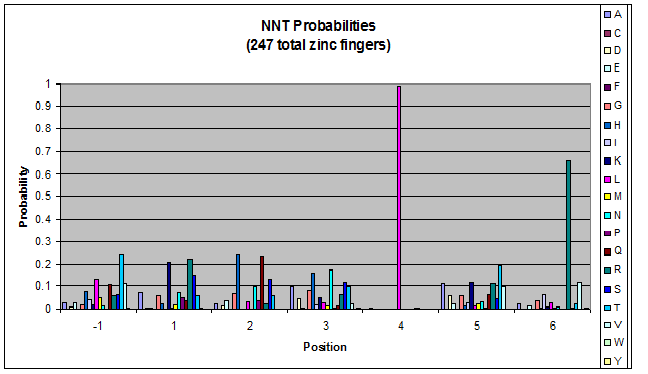
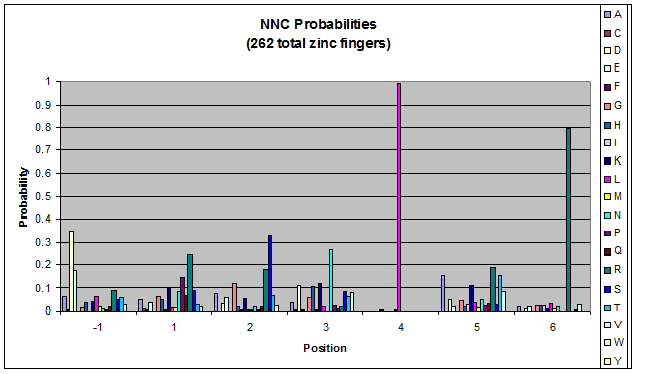
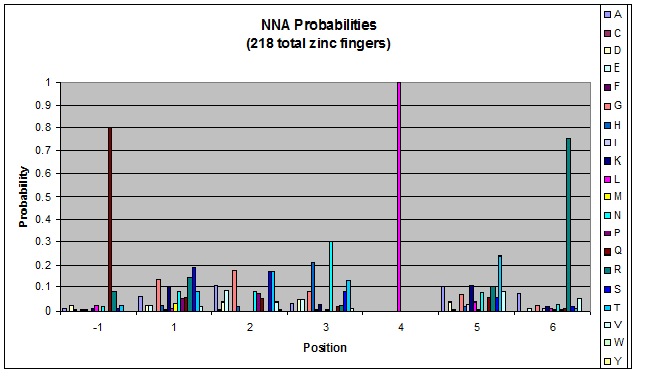
Probability data
- The following are graphs of the probability of finding each amino acid at each position on the alpha helix.
June 21st
Wet Lab
His3 sequencing results:
The sequencing results showed that the His3 (HisB) gene is still present in the strain and without any early stop codons. There is a 2 aa deletion in the middle of the protein, but its purpose is unknown and the gene likely is still fully functional.
- Restreak selection strain on plate from glycerol stock--tomorrow we will PCR the His3 locus and sequence again just to be sure.
- Made oligos for MAGE to insert stop codons and make a frame shift in the endogenous His3 gene, so that if necessary we can knockout His3 ourselves.
Selection strain with lambda red:
- Reinoculated and made glycerol stock
- prepared for MAGE tomorrow
June 21st - Bioinformatics
Persikov Statistics - Graphs
- FQCRICMRNFSzif268 F2 Backbone/Helix F1/TGEKPlinker
- The Persikov data shows weak predictive power for OPEN amino acid sequences. Our conclusion is that Persikov's program is not well-suited for incorporation into our helix generator. Testing Persikov's helices in his program yeilded mostly accurate results (approximately 24/25 matched known binding information). This is an important test because it proved that we are using the program correctly and that the program is in fact working properly. However, testing the OPEN sequences in Persikov's program resulted in numerous false negative values which informed our decision not to use Persikov's program to check our own hellix-generating program.
Phone Call with Dan
- How conservative/risky should we be in terms of using other backbones?
- Conservative
- Possible Pros:
- More likely to get something that will work
- Depending on how "smart" our probabilities are (from our ZF generation algorithm), we could cover a lot of novel space without straying too far from zif268
- Worst Case:Something we can show for iGEM (we covered the same ground OPEN did, and found many of the same ZFs, but with a targeted approach, a "smarter" method-- not throwing random things at it; Chip is not ours, but the program is "smarter")
- Possible Cons:
- Might end up covering the same ground as OPEN, but doing a "worse" job than they did
- Less likely to discover new/groundbreaking things (i.e., TNN triplets)
- Possible Pros:
- Less Conservative
- Have 3-6 target sequences (we're currently going for 8)
- More backbones from non-zif268 than zif268
- Pros:
- We could get luck and find something no one has ever seen before (TNN, ANN). If we throw enough things at it, we're more likely to get luck.
- Cons:
- Risk: Many of these backbones (from entire ZF world)may NOT bind DNA (i.e., may bind proteins)
- Risk: May not find anything that binds, then the whole project is a dud
- Conservative
- What is the more important variable, helices or backbones?
- Helices seem to be more important, backbones of secondary importance
- Backbones: ZF's unravel DNA, open the major groove-- backbone is important here, changes the bond angle, etc. (Brandon's paper-??)
- Balance needed between low and high risk
- If we find backbones that we know bind DNA, greatly lowers our risk
- Limited spaces on chip: zero-sum game
- With a middle of the road approach, we diminish both benefits and risk (diminishes the benefits of the high risk approach much more than it diminishes the benefits of the conservative approach; i.e., if you're playing the lottery, you're more likely to win if you buy many more tickets)
- We need to compare probabilities of randomly-generated OPEN sequences vs. probabilities of sequences randomly generated by our program
- OPEN tries to cover all space: smaller probability
- If we have a "smarter" algorithm, we can produce fewer
- However, the idea is not to repeat OPEN, but to go somewhere else, non-GNN sequences
- Remember: OPEN is a Cell paper; the point of the project is not to compare ourselves to them
- If we find binders for 1-2 of our sequences, that would be awesome
- Probably we'll have some that find none, some have 10, our last one might have 1,000 hits (then, we do bioinformatics to figure out why/what those hits were)
- Point: to learn and do high-level bioinformatics, and high-tech cloning techniques in the lab
- If you do find binders, you can write a paper about it!
- We have all the resources we need right now to build our chip
- We need to pick out targets
- Need to decide exactly what we want for:
- No. of target sequences/which ones
- No. of helices/ which ones
- Ratio of zif268 backbones: non-zif268 backbones
- Avoid switching Leucine out of position 4, then change other positions based on our frequencies
Chip Design
- No. of sequences will be more than we can put on the chip
- Helices: essentially unlimited
- Put more-likely-to-bind helices into the risky backbones
- Put less-likely-to-bind helices into a zif268 backbone
- Helices: essentially unlimited
- Backbones
- Maybe revert to a more targeted approach: pick backbones that we know are transcription factors (TFs), that we know bind to DNA
- OR research the ZFs from the phylogenetic tree
- Pick clades to research, see if one looks better than the other
- Why did OPEN cover so many helices, without changing the backbone, but still yield predominantly GNNs?
- If we have an idea of how the backbone might affect binding, maybe we could look into some sort of low-level modeling, etc. so that we wouldn't be grasping? Could Vatsan help with this?
- See 2000 Wolfe paper [http://www.ncbi.nlm.nih.gov/pubmed/10940247]
- Backbones could affect interactions between fingers
- Theory: energy penalty to ZF binding-- unravels DNA when binds to it
- We have 12 target sequences
- 2 per 4 diseases, 4 for the 5th disease
- If we want to be more conservative, we could throw out Type III, but it could be something cool
- We should have mostly Type I (CoDA argument, if this is an F2)
- Proposed: 3 diseases, 6 sequences
- 4 Type I (F3 and F2 known, F1 novel)
- 1 Type II (GNN, ANN, GNN)
- 1 Type III (All unknown, e.g., TNN, ANN, TNN;max 1)
Or, for 3 diseases:
- Type I's
- Type I, Type II
- Type I, Type III
- Clinical Targets
- Colorblindness (Type I's)
- Familial Hypercholesterolemia (FH) (1 in 500)
-
Cystic Fibrosis (CF) -
Tay Sachs - KRAS- (oncogene/cancer)
- Main goal of project: to build outside of what is already known
- If we wanted to cure a disease only, we could just use existing ZFs (i.e., find GNN binding locations)
- Also, we lend a level of specificity for insertion/deletion
- There is the possibility that there might be some area where specificity might demand ANN codons
Current decision on chip design:
- We will have 6 target sequences, 2 each from colorblindness, FH, and KRAS. All are "Type I" targets (only F1 is novel) with the middle finger chosen from the CODA paper (either GNN or TNN)
- N.B.: the CB and FH sequences make up full ZF nuclease cut sites. The KRAS sites, due to the small number of GNNTNN F3F2 combos available in CODA, are separate, with the flanking ZF nuclease site added afterwards in parentheses
- GGTGGTAAG (CB)
- GGAGTCCTG (FH)
- GGCTGATGC (KRAS) (CTGAAAATT)
- GGCTGACAC (FH)
- GGCTGGAAT (KRAS) (GACAAGAGC)
- GTCGCCTCC (CB)
- Targets 3, 4, and 6 are similar to sequences Zif268 variants successfully bind to, so the backbones will be weighted accordingly:
- Zif268_F2 backbone: 6000 helices (per target)
- 10 backbones more closely related to Zif268: 300 helices each
- Targets 1, 2, and 5 will have equal distributions of backbones:
- Zif268_F2: 3000 helices
- 10 backbones closely related to Zif268: 300 each
- 10 backbones more distantly related to Zif268: 300 each
Identifying dependencies
- We looked at the probability graphs to determine which amino acid positions on the finger's helix interact with which bases.
- Some interactions are fairly well estabilished, while others have been more recently proposed (See interaction map (Persikov 2011))
- To identify these interactions in our own data we looked at which helix positions varied most when you changed the bases. A more rigorous way to do this is to calculate the entropy change as you change the amino acids in each position.
- xNN(Vary base 1): Amino acid 6 changes
- NxN(Vary base 2): Amino acid 3 changes
- NNx(Vary base 3): Amino acid -1 and 2(?) changes
- Our program looks at dependencies between amino acids when generating sequences.
- We decided on these amino acid dependencies, using both established data and patterns we saw in the OPEN data:
- -1 and 2
- 2 and 1
- 6 and 5
- We decided on these amino acid dependencies, using both established data and patterns we saw in the OPEN data:
- Because there is not much data for 'CNN' and 'ANN' sequences (with 16 and 29 known fingers that bind to each triplet, respectively), we should use pseudocounts for these sequences, so that our frequency generator is not too biased toward probabilities that may not be significant.
June 22nd - Wet Lab
Preparing media/reagents for selection system:
- Made 0.1M zinc chloride solution, M9 salt, and 1M magnesium sulphate solutions for the amino acid mixture
- M9 Salt solution (20x)
- 67.8 g of disodium phosphate
- 30 g of monopotassium phosphate
- 5 g of sodium chloride
- 10 g of ammonium chloride
- All in 500 mL of distilled water
- Sterile filtered when done dissolving
- M9 Salt solution (20x)
Overhang PCR for 3-part assembly of ZFs, omega subunit, and backbone vector (pZE21G, spec resistance)
- Clone out omega+Zif268:
- template: original selection construct plasmid (ZFB, his3, etc.)
- primers: omega_F+homolog, Zif268_R+homolog
- Protocol
- 98 C for 30 sec
- 98 C for 10 sec
- 68 C for 30 sec
- 72 C for 30 sec
- Repeat steps 2-4, 30 times
- 4 C for ever
- Clone out omega only:
- template: original selection construct
- primers: omega_F+homolog, omega_R
- Protocol
- Same as omega + Zif268
- Clone OZ052 with overhang:
- template: OZ052 overhang (overhang currently matches selection construct), 1:10b, 15ng/µL
- primers: OZ052_F+omega homolog, OZ052_R+homolog
- Protocol
- Same as omega + Zif268
- Clone OZ123 with overhang:
- template: OZ123 overhang (overhang currently matches selection construct), 1, 6.3ng/µL
- primers: OZ123_F+omega homolog, OZ123_R+homlog
- Protocol
- Same as omega + Zif268
- clone out pZE21G backbone
- template: pZE21G containing cells from plate, diluted 1:10
- primers: back_F, back_R
- Protocol
- 98 C for 30 sec
- 98 C for 10 sec
- 68 C for 30 sec
- 72 C for 1:30
- Repeat steps 2-4, 30 times
- 72 C for 5 min
- 4 C for ever
- 25µL reaction:
- 12.5µL Phusion mastermix
- 1.25µL each primer
- 1µL of 1ng/µL dilution of template
- 9µL of ddH2O
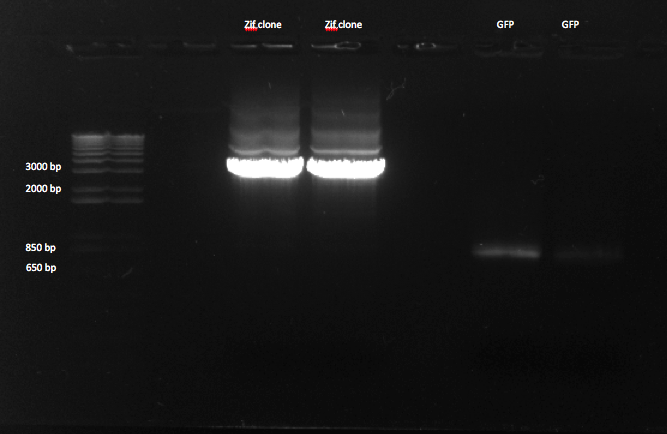
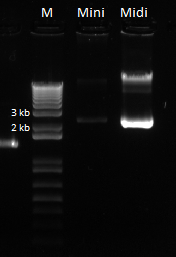
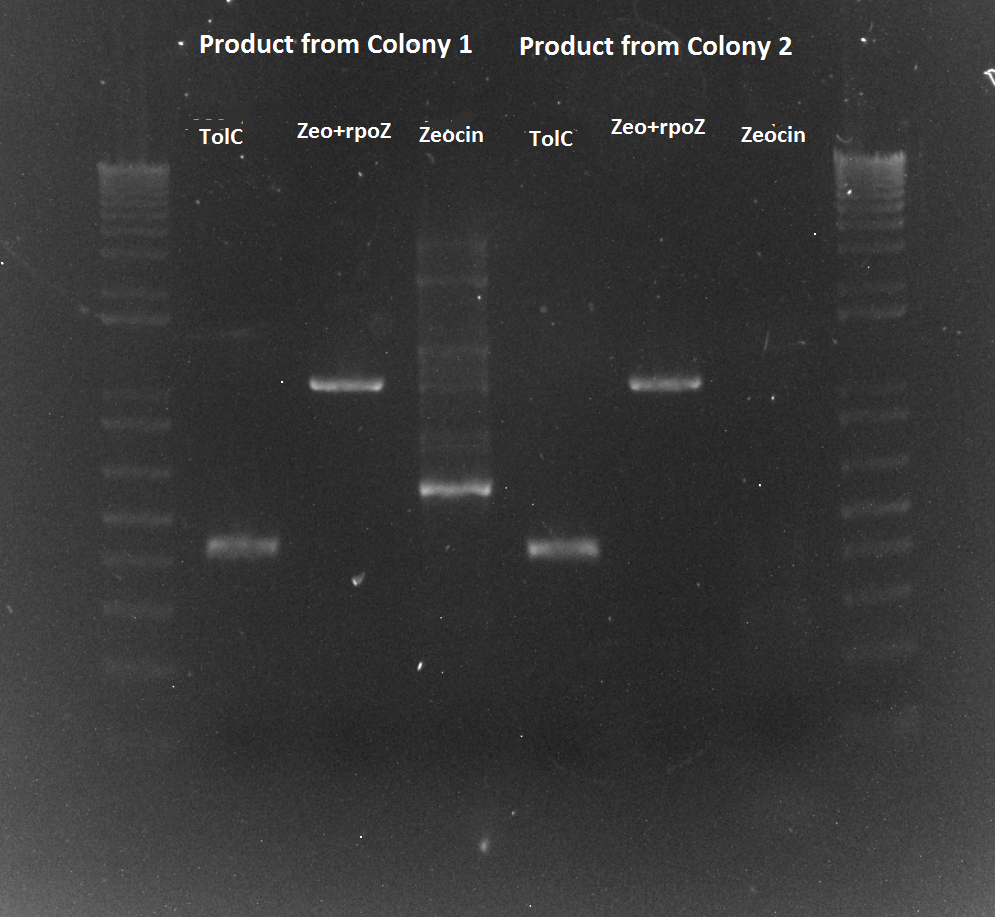
- Ran gel of the above PCR products and imaged below: only the omega and omega+Zif268 reactions seemed to work
pZE21G backbone:
- 1 colony of pZE21G grown in 3mL LB, 3µL spectinomycin (1000x) until mid-log. Glycerol stock made.
- Miniprep of pZE21G in order to PCR the backbone: used Qiagen kit
- 5ng/µL and didn't seem pure: miniprep (or nanodrop) not working
- another colony used to start an overnight culture for PCR/glycerol stocks
- Ran gradient PCR on the miniprep product in order to obtain backbone
- same protocol as before, with 1µL of template
- 6 tubes spaced so that annealing temp=60,62,64,66,68,70
- Parameters: (program on PCR5, IGEM-> DOGGED)
- 98°C for 30s (initial denaturation)
- 98°C for 10s (denature)
- 60°C to 71°C for 15s (anneal)
- 72°C for 90s (extend)
- Repeat steps 2-4 for 30 cycles total (denature, anneal, extend)
- 72°C for 5 min
- 4°C forever
HisB locus PCR: We repeated the PCR of the selection strain at this locus just to be sure HisB is still present
- grew 1 colony from a new selection strain plate Vatsan brought in 0.5mL LB, 0.5µL tet
- used 1 µL of bacterial suspension for PCR following same procedure as 6/20
Lambda red to make selection system:
- grew ?His3?PyrF?rpoZ+pKD46 to mid-log (0.4 using OD)
- induced lambda red by shaking culture in 42C water bath for 15 min
- spin down 1mL for 1 min, 18000 rcf at 4C
- wash 2x with cold water, removing as much supernatant as possible
- resuspend with 200 ng kan-ZFB-wp-his-ura template (20µL) and water up to 50µL (30µL)
- electroporate using 1mm gap cuvettes adn 1.80KV. Immediately afterward add 1mL LB to cuvette, mix, and transfer to culture tube containing 2 mL more of LB
- recover for 2hrs, 30C
- spread on kanamycin plates: 100µL, 10µL, or 1µL (the last two dilute with 100µL LB to help spread more easily)
- grow overnight at 30C
June 22nd - Bioinformatics
Final target sequences
Our "tentatively" Final DNA Target Sequences (i.e. barring any major objections, we're going with this):
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,402,679-153,408,753 | 256 | GGC TGA GGC | GTA GCT GGG | ESGHLKR.QREHLTT.####### | QSGTLTR.QRSDLTR.KKDHLHR |
| Colorblindness | chrX:153,402,679-153,408,753 | 2067 | GAA GGG GAC | GGG GCT CAC | QDGNLGR.RREHLVR.EEANLRR | RTEHLAR.QRSDLTR.####### |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 2707 | GGC TGG ATG | GGC TGG CTC* | ESKHLTR.RREHLTI.####### | ESKHLTR.RREHLTI.####### |
| Pancreatic Cancer | chr7:117,074,084-117,089,556 | 4423 | GCA GAC TGT | GCA GGA AAA | QGNTLTR.DRGNLTR.####### | QDVSLVR.QSAHLKR.####### |
- Drier was unable to find a ZF that bound specifically to CTC. Instead he found zinc fingers that bound to CTC and other sequences with equal binding affinity.
- Note: The green cells are the target sequences that we are aiming for on our chip.
Finalizing the non-Zif268 backbones
In addition, we locked down the non-Zif268 backbones that we will be using for the chip. We have 10 backbones that are more closely related to Zif268, and 10 that are more distantly related:
| More Closely Related Backbones | More Distantly Related Backbones | |||
| Name | Sequence (with helix) | Name | Sequence (with helix) | |
| 44GLAS_DROME | FRCPI---CDRRFSQSSSVTTH-MRTH-- | 56EGR1_HUMAN | FAC---DICGRKFARSDERKRHTKIH--- | |
| 38KRUP_DROME | FTCKI---CSRSFGYKHVLQNH-ERTH-- | 47MZF1_HUMAN | FVC---GDCGQGFVRSARLEEHRRVH--- | |
| 124EVI1_HUMAN | YRC---KYCDRSFSISSNLQRHVRNIH-- | 23CF2_DROME | YTC---SYCGKSFTQSNTLKQHTRIH--- | |
| 6HUNB_DROME | YECK---YCDIFFKDAVLYTIHMGY--H- | 19ZEP2_RAT | YICE---ECGIRCKKPSMLKKHIRTH--- | |
| 16SUHW_DROME | FPCEQ---CDEKFKTEKQLERH-VKTH-- | 49SDC1_CAEEL | VVC---FHCG-TRCHYTLLHDHLDYCH-- | |
| 125CF2_DROME | YTC---PYCDKRFTQRSALTVHTTKLH-- | 27SDC1_CAEEL | LTC---AHCDWSFDNVMKLVRH-RGVH-- | |
| 43EVI1_HUMAN | FKCHL---CDRCFGQQTNLDRH-LKKH-- | 130TTKB_DROME | YRC---KVCSRVYTHISNFCRHYVTSH-- | |
| 118ADR1_YEAST | YPC---GLCNRCFTRRDLLIRHAQKIH-- | 80ESCA_DROME | YQC---PDCQKSYSTFSGLTKH-QQFH-- | |
| 24EVI1_HUMAN | QECK---ECDQVFPDLQSLEKHMLS--H- | 20IKZF1_MOUSE | HKCG---YCGRSYKQRSSLEEHKERCH-- | |
| 25SUHW_DROME | MSCKV---CDRVFYRLDNLRSH-LKQH-- | 127SRYD_DROME | QECTT---CGKVYNSWYQLQKHISEEH-- | |
Updated Chip Design
The CODA article produced zinc fingers that bound a GNN or TNN F2 with either a ANN, GNN, or TNN F3. These results lead us to the following distribution of three types of zinc finger backbones (Zif268, similar but not equal to Zif268, and dissimilar to Zif268) across our 6 target DNA sequences. With 55,000 spaces on our chip, each of the 6 target DNA sequences is allotted 9,150 spaces with 100 spaces set aside for control zinc fingers from CODA and OPEN. Note that the values in the table below represent the number of helices inserted into each type of backbone.
| Disease | Target DNA Finger 2 | Target DNA Finger 1 | Helices in Zif268 Backbone | Helices in Zif268 Closely-Related Backbones | Helices in Zif268 Distantly-Related Backbones |
| Colorblindness | TNN | GNN | 5150 | 3000 | 1000 |
| Colorblindness | GNN | CNN | 3050 | 3050 | 3050 |
| Familial Hypercholesterolemia | TNN | ANN | 3050 | 3050 | 3050 |
| Familial Hypercholesterolemia | TNN | CNN | 3050 | 3050 | 3050 |
| Pancreatic Cancer | GNN | TNN | 5150 | 3000 | 1000 |
| Pancreatic Cancer | GNN | ANN | 3050 | 3050 | 3050 |
N.B.: The chip will only be holding our F1 zinc fingers- the F2 and F3 will be on a separate plasmid that we must make ourselves
To Do: The distribution of helices to each backbone set/target sequence needs to be finalized. For example, the program can generate a set of helices for the Zif268 backbone to be applied to the colorblindness target sequence, but should the same set or a completely different helix set be applied to the Zif268 backbones for the familial hypercholesterolemia target sequences?
- If we want to test the effect of the backbone, would need to keep the helices constant-- but we could do this within a single target, to keep all other variables constant
Finishing the generator
We finished and finalized the program that generates zinc finger sequences. The following changes were made today:
- We incorporated the data from Persikov's database into out generator.
- We included the ability to remove duplicate sequences in the output file (with a dictionary).
- We added pseudocounts for fingers that bind to 'ANN' or 'CNN' targets. Because there is not much information for these targets, the data we do have may be biased. Thus, we want to make sure that amino acids that currently have no probability of occurring are bumped up to a minimum (currently 0.01).
- We placed additional weight on non zif-268 backbones. Formerly, the amino acids for positions 1, 4, and 5 were fixed based on zif-268 data, regardless of the original helix sequences on these backbones. Now information from both zif-268 and the original helix sequence is considered when assigning weights to the amino acids.
- We started working with Noah's reverse translate program.
We tested our generator to ensure that the sequences it was producing appeared to be legitimate.
- Jamie looked at the multiple sequence alignments of the fingers generated, so see if the frequencies correlated with what we expected them to be. (?)
- We input a known DNA triplet to see if the program generated sequences known to bind, according to OPEN data. When generating 10000 sequences, about half the known binders for the input triplet were found.
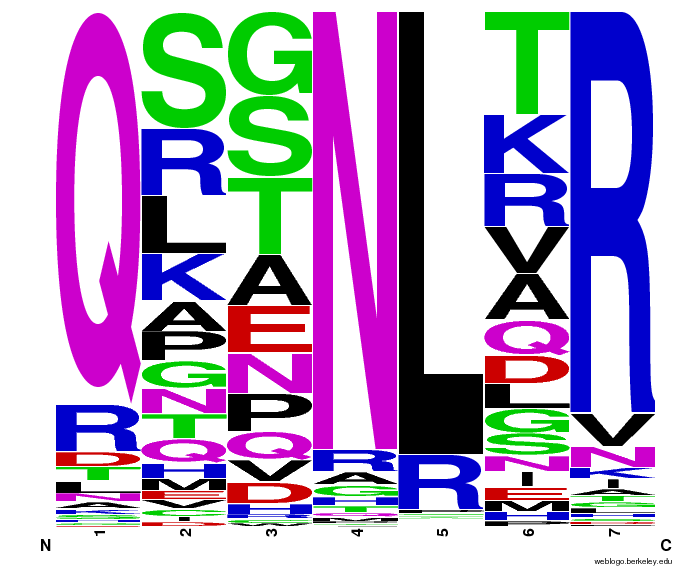
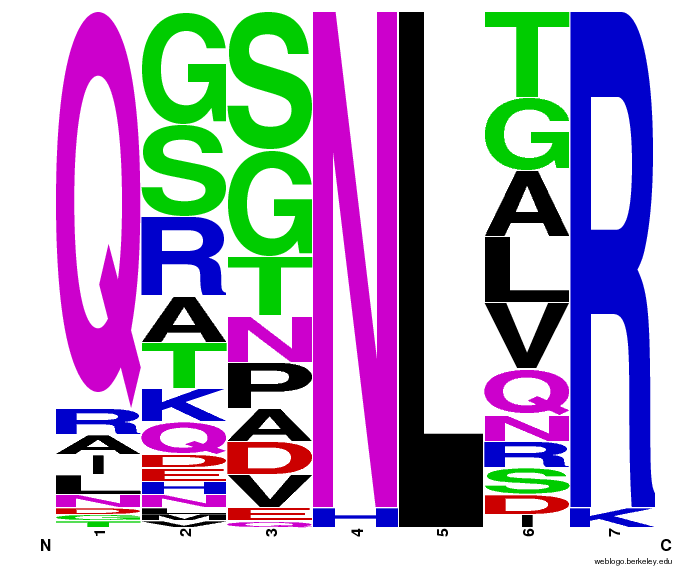
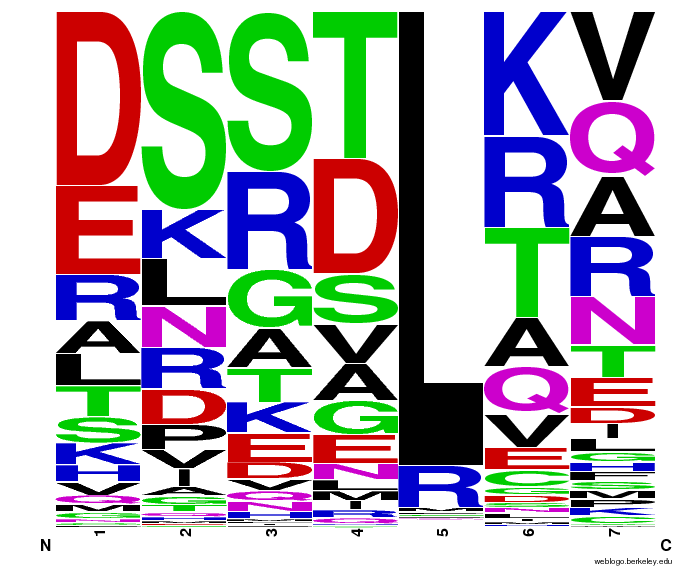
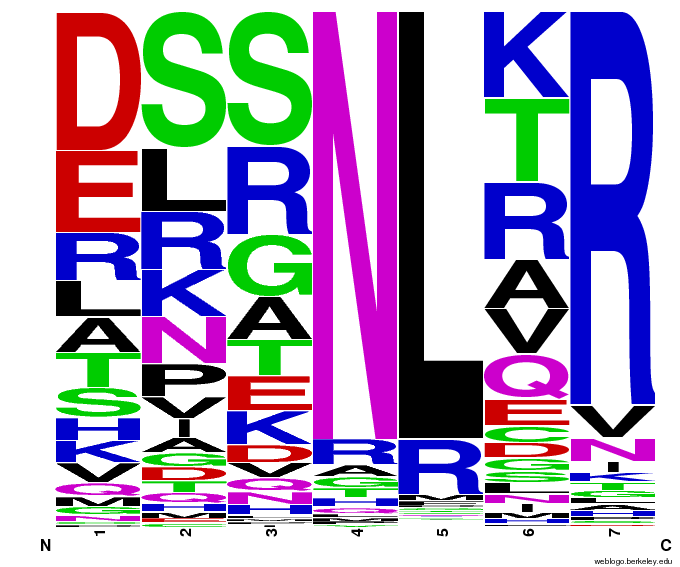
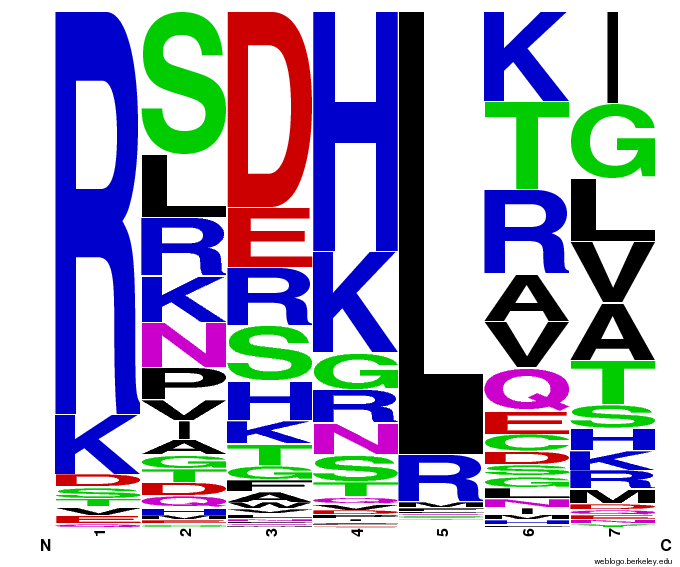
We created [http://weblogo.berkeley.edu/ WebLogos] to more easily visualize how adding the Persikov data affects the sequences we generate. The size of the letters correspond to the frequency of that amino acid in that position. We decided to incorporate the Persikov data so that our generator incorporates more information when generating sequences. Doing so does not drastically change the sequences generated.
June 23rd - Wet Lab
Ran gel to determine the results of the PCR products
- Determined the HisB presence in selection strain
- Finalized the presence of hisB through the gel image below
- Determined the success of the pZE21G backbone primers through gel on gradient PCR
- PCR for the backbone failed again, even done through gradient PCR
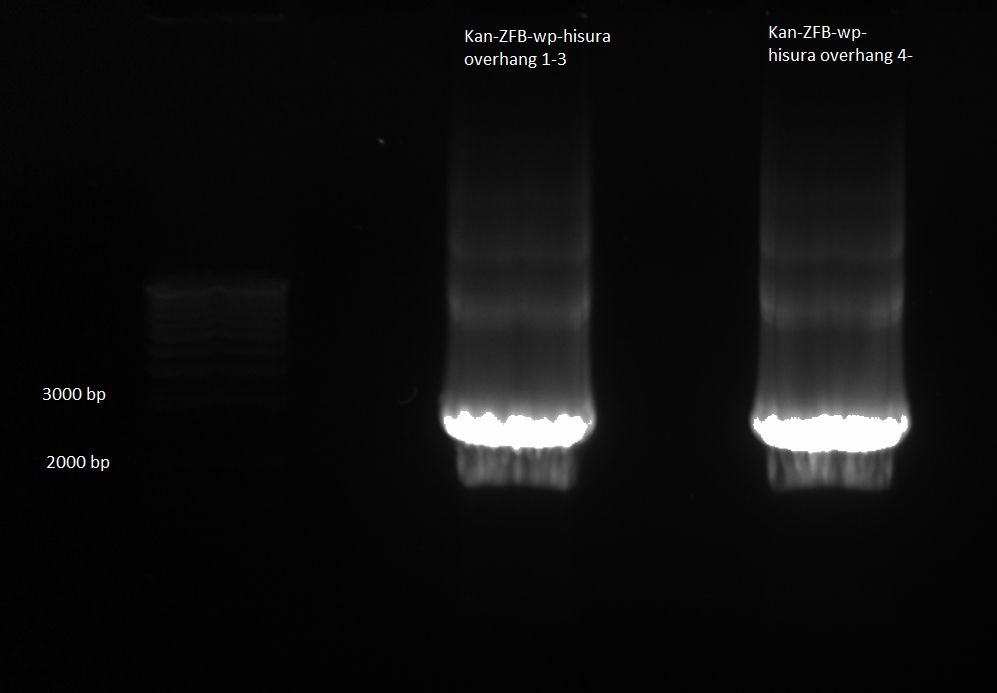
PCR more Kan-ZFB-His3-Ura3
- Two 50 µL reactions
- Doubled the protocol used on the 16th and used HisUraKan_F and ZFBwpHisUra_R primers
- PCR produced very low concentration of Kan-ZFG-His3-Ura3 because melting temperature of primer was too high, so primers stuck to annealing DNA and did not dissociate
- In image below the low concentration of desired product can be seen, along with the high concentration of unused primers
- Ran gel of initial overlap PCR product (undiluted, starting without primers) from June 16th, using whole sample
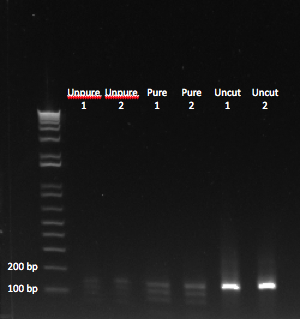
- Performed gel extraction in order to have more Kan-ZFB-His3-Ura3 product for the transformation tomorrow: 8.5ng/µL, 260/280=2.03
Determined success of the selection construct transformation
- Checked the plates all day,and finally came to the conclusion that the transformation did not work
- Discovered that lambda red has a promoter induced by arabinose, not temperature (though the strain is still temperature sensitive). That is why it didn't work--we'll get arabinose and hopefully have a successful recombination.
- Preparing all parts for transformation today and will finish it tomorrow
Oligo Design for MAGE
- Designed 90bp long oligos for OZ052 and OZ123 insertion in the ZFB sites in place of Zif268. Reverse complement taken.
Miniprep pZE21G plasmid for backbone PCR
- Ran miniprep again for pZE21G plasmid: 6ng/µL, 260/280=2
- Worried that the miniprep didn't work: ran gel also on this miniprep and concluded that DNA was present in the sample
- Gel image seen below
Ran PCR for pZE21G backbone
- Used same protocol from June 22 in PCR today for pZE21G backbone
June 23rd - Bioinformatics
Revising Target Sequences
| Target DNA | Cystic Fibrosis | Familial Hypercholesterolemia | Retinal Blastoma | p53 | Myc | Pancreatic Cancer |
| GNN A | Flank 1 | ? | ||||
| GNN T | Flank 1 | |||||
| GNN C | ? | Flank 2 | ||||
| TNN G | Flank 2 | X | ||||
| TNN C | Flank 3 | ? | ||||
| TNN A | Flank 3 | ? |
This is the set of final, final target sequences based on the table above:
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,403,001-153,407,000 | 3627 | GCT GGC TGG | GCG GTA ATG | EGSGLKR.EAHHLSR.####### | RRDDLTR.QRSSLVR.####### |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 14001 | GGC TGA GAC | GGA GTC CTG | ESGHLKR.QREHLTT.####### | QTTHLSR.DHSSLKR.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 198 | GGT GCA GGG | GGC TGA CTC | VDHHLRR.QSTTLKR.RRAHLQN | ESGHLKR.QREHLTT.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 981 | GGA GAG GGT | GGC TGG AAA | QANHLSR.RQDNLGR.TRQKLET | EKSHLTR.RREHLTI.####### |
- Green cells are our target sequences.
June 24
- Designed primer for testing HisB deletion, reuse His_Internal_R to test the band
pZE21G:
- reinoculated culture with 100µL of saturated solution, grew to mid-log, and made glycerol stock
- backbone PCR: ran E gel but no bands--PCR unsuccessful. We may need to use a different backbone for the zinc fingers.
Omega and Omega+Zif268:
- these were the only two PCR reactions from 6/22/11 to work
- PCR purified using Qiagen kit:
- omega: 6.1ng/µL, 260/280=1.83
- omega+Zif268: 11.3 ng/µL, 260/280=1.67
Lambda red recombination of selection system:
- reinoculated selection strain+pKD46 with 100µL of saturated solution
- just before mid-log (about 4 hours after inoculation) divided culture in half (1.5mL) and added either 37.5µL or 3.75µL of 20% arabinose solution (to try two different induction levels). Cultures grew for another hour.
- The rest of the procedure was the same as the 6/22/11 attempt but without the 42C water bath.
June 24th - Bioinformatics
Playing with Pseudocounts
Using CTC because of position 6's reliance on the CNN frequencies, we see what difference values of pseudocounts (if in the frequency table, the frequency of an amino acid is 0, bump it up to the psuedocount: ex. A = 0 becomes A = .015 with a psuedocount of .015) make. Pseudocounts are necessary for data that has small sample size - we could be missing out on working helices because a letter's frequency is 0 when it shouldn't be.
Various pseudocount (psu = ) values. Look at the 7th column, which is position 6 in the helix:
The variation from E being the top letter to A being top back to E is from a slight adjustment in how we add on psuedocounts: the 'new' way is a more proportional approach.
Notice how psu = 0 gives only the four letters found in our dataset, while psu > 0 adds in other letters, each with a small probability ranging from .5% to 2%.
The question is how much psu to add: less means we weight our (possibly flawed) data of proven zinc fingers more. Higher psu adds more randomness (variation) to our sequences, but some fraction of those sequences will not work.
Updated Closest Zif268 Fingers
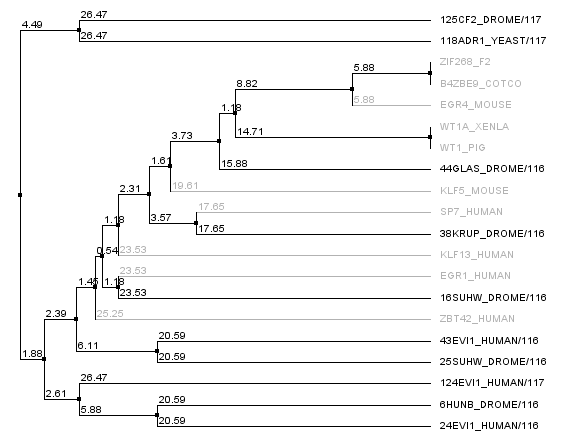
We realized that some of our "close non-zif268 fingers" were actually not all that close to Zif268, and so we went into the 88,000 zinc finger database and pulled out zinc fingers surrounding zif268. In fact, there were many, many, many zinc fingers that had identical sequences to the Zif268 F2 finger, and so we looked at sequences around it. The tree below shows the new non-zif268 backbones that are actually close to zif268 compared to our old set. The new set is in gray, the old set is in black. This gives us a potential seven more backbones to work with.
Sequence Generation
We made some small updates to the sequence generator, based on the frequencies we noticed in the outputs of the tests we ran.
- We decided to only include pseudocounts for position 6 for 'CNN' and 'ANN.' Originally, 'CNN' and 'ANN' were using pseudocounts for all seven positions. However, this introduced a noticeable increase in amino acids, such as tyrosine (Y), that have been shown to occur rarely in zinc fingers (according to our data from OPEN and Persikov). Additionally, because tryosines occured so rarely in the data (11 times total in the open data set), we decided not to give tyrosine a pseudocount.
- We added the capability to prevent repeat backbone-helix combinations on the chip. That is, we wanted to make sure that the same exact zinc finger was not generated for different triplet inputs.
To test the sequence generator, we made two sets of 2000 sequences for GAA, then infographic-d the results. Comparing these with the images for OPEN and OPEN+Persikov shows that our generation follows the major themes of those datasets, but also introduces variation. The two generated sets also vary slightly from each other, which shows the influence of randomness on the generation.
| Disease | Target DNA Finger 1 | Helices in Zif268 Backbone | Helices in Zif268 Closely-Related Backbones | Helices in Zif268 Distantly-Related Backbones |
| Colorblindness (Bottom) | TGG | 5150 | 3000 | 1000 |
| Colorblindness (Top) | ATG | 3050 | 3050 | 3050 |
| Familial Hypercholesterolemia (Bottom) | GAC | 5150 | 3000 | 1000 |
| Familial Hypercholesterolemia (Top) | CTG | 3050 | 3050 | 3050 |
| Myc (Top198) | CTC | 3050 | 3050 | 3050 |
| Myc (Top981) | AAA | 3050 | 3050 | 3050 |
Table of target sequences and helix distribution across backbones
- Distribution: Zif268 : Zif268 similar : Zif 268 dissimilar
- Conservative distribution 56.3 : 32.8 : 10.9
- Riskier distribution 33.3 : 33.3 : 33.3
List of Remaining Goals:
- Sort fingers by target
- Pick and assign primer sets
- Reverse translate fingers avoiding type II restriction enzymes and primers
- Append type II restriction enzyme and primer sequences to each finger
- Yay
June 25th-26th - Bioinformatics
This is the set of final target sequences with assigned forward and reverse primers (tags for PCR):
| Disease | Target Sequence | Forward Primer (5'-3' NOT REVERSE COMPLEMENT) | Reverse Primer (5'-3' NOT REVERSE COMPLEMENT) |
| Colorblindness | GCT GGC TGG | ATATAGATGCCGTCCTAGCG | AAGTATCTTTCCTGTGCCCA |
| Colorblindness | GCG GTA ATG | CCCTTTAATCAGATGCGTCG | TGGTAGTAATAAGGGCGACC |
| Familial Hypercholesterolemia | GGC TGA GAC | TTGGTCATGTGCTTTTCGTT | AGGGGTATCGGATACTCAGA |
| Familial Hypercholesterolemia | GGA GTC CTG | GGGTGGGTAAATGGTAATGC | ATCGATTCCCCGGATATAGC |
| Myc-gene Cancer | GGC TGA CTC | TCCGACGGGGAGTATATACT | TACTAACTGCTTCAGGCCAA |
| Myc-gene Cancer | GGC TGG AAA | CATGTTTAGGAACGCTACCG | AATAATCTCCGTTCCCTCCC |
June 27
Wet lab
Sequencing PyrF, rpoZ loci:
- We will sequence these genes in the selection strain just to make sure they are knocked out, especially since it appears HisB is not.
- Picked a colony off ∆HisB∆PyrF∆rpoZ plate (6/21) and grew in 150µL LB plus tet in a 96 well plate for about 2 hrs at 37˚C
- diluted 1 in 20 and used 1µL as template in PCR with KAPA mastermix (see protocols for reagent amounts and parameters)
- annealing temp 65˚C, elongation time 1:15
- PyrF_F, PyrF_R primers
- PyrF_F, PyrF_internalR
- rpoZ_F, rpoZ_R
- rpoZ_F, rpoZ_internalR
- rpoZ_R, zeocin_R
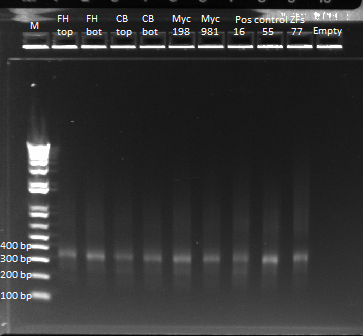
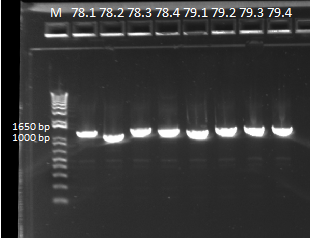
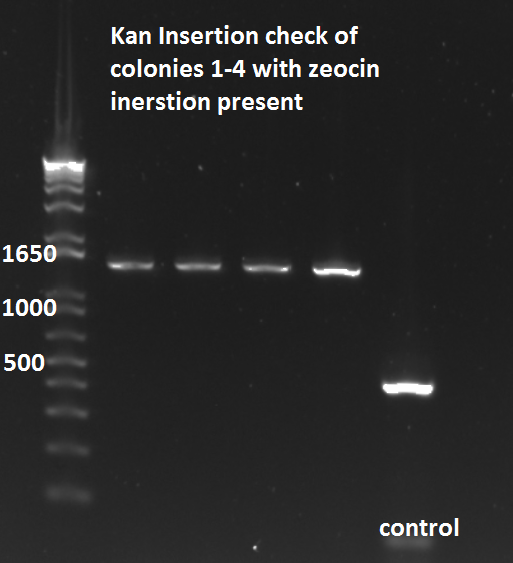
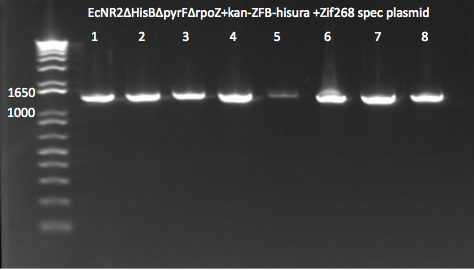
- Run on E Gel to check PCR worked: bands are at the same sizes as the original genotyping gel.
- Tomorrow we will send samples to Genewiz for sequencing
Lambda Red recombination:
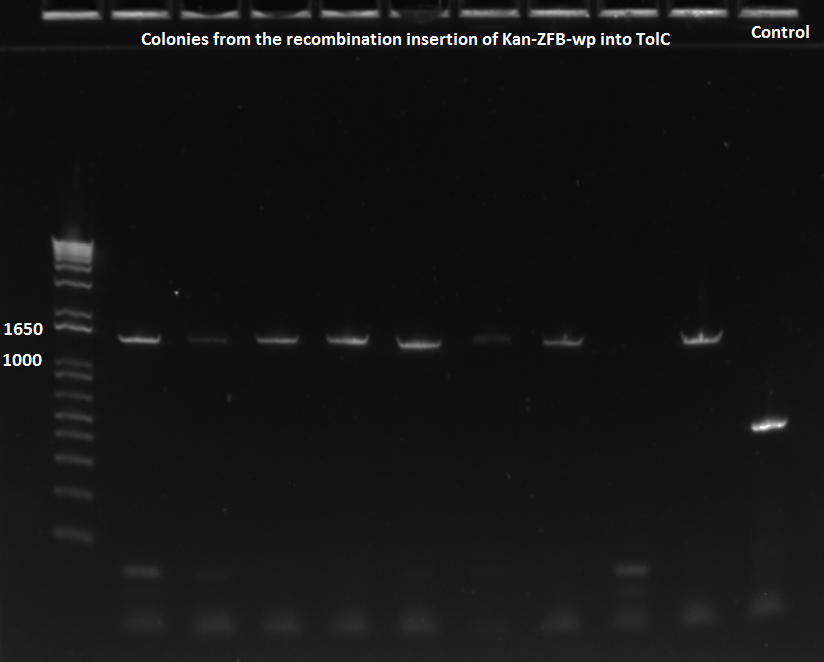
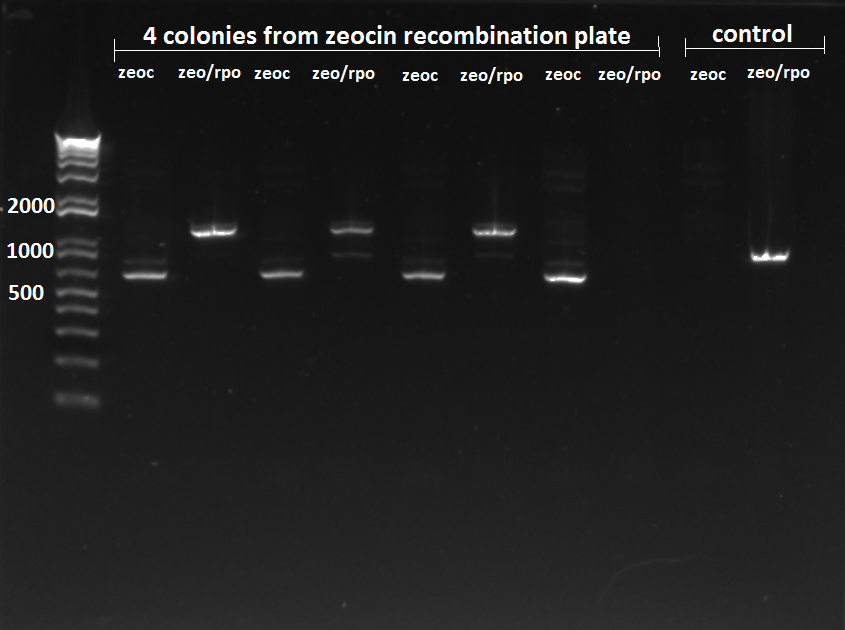
- The plates made from the recombination (6/24) did have colonies, but they were very small and took a long time to grow, and so they may not actually have the kan-ZFB insert. We will have to PCR the locus to see.
- Chose 8 colonies from each plate and grew at 30˚C in 150µL LB plus kan in a 96 well plate
- When our primers arrive, we will PCR the locus to check for the insert.
Selection system media:
June 27th - Bioinformatics
To Do for Today
- 100 sequences (and control), 2 each with the same F3 and F2, but different F1, from our test sequences [zif268, OZ123, OZ052, CoDA]✓
- Type II nuclease cut site sequences- put the binding sites into our oligos ✓
- Final backbones with helices ✓
- Programming stuff- Check to make sure there are no cut sites or primers in any of our backbone/helices combinations; check translation order (translates F1→F3)✓
100 Control Sequences
- See our Positive Control Sequences, updated June 28th
- Selected known binding zinc fingers from the CODA table that bind sequences similar to our target sequences
- All control helices from CODA were inserted into Zif268 F2 backbones and have been assigned a seventh primer tag separate from the tags given to the 6 target sequences.
Updated Target Sequences
One of our sequences from before was bad because the F3/F2 combo did not appear in the CODA table... faulty checking, my bad :(
Here is the newest table of target sequences:
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,403,001-153,407,000 | 3666 | GTG GGA TGG | GAA GGG ACC | RNTALQH.QSAHLKR.####### | QDGNLGR.RREHLVR.####### |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 14001 | GGC TGA GAC | GGA GTC CTG | ESGHLKR.QREHLTT.####### | QTTHLSR.DHSSLKR.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 198 | GGT GCA GGG | GGC TGA CTC | VDHHLRR.QSTTLKR.RRAHLQN | ESGHLKR.QREHLTT.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 981 | GGA GAG GGT | GGC TGG AAA | QANHLSR.RQDNLGR.TRQKLET | EKSHLTR.RREHLTI.####### |
- Green cells are our target sequences.
Cut Site Design
- See our Cut Site Design page
- We left in one proline (P) between the linker and the starting FCQ... of finger 2, but as this proline is the last AA of the OPEN linker (TGEKP) and occurs before the beta sheet in every zinc finger in Zif268 (see zif268's sequence on its [http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=1AAY PDB page])
- This configuration also allows the library to be used at any finger position because proline ends the OPEN linker.
Updates on the program
The program appears to run extremely slowly because of the computationally intensive step of checking the reverse translated sequences
- In addition to checking for the primers and cutsites, we also have to check for 'GGGGGG' because it can lead to undesirable structures forming. In addition, we have to check for the reverse complements for all these undesirable sequences.
- We decided on a similarity of 0.8 as the maximum acceptable similarity between the sequence the primer bind to and any other part of the generated sequence. If the sequences are too similar, the primer might mishybridize. We originally had a similarity threshold of 0.6 but that made the program run too slowly, so we decided on a threshold of 0.8.
June 28th
Sequencing:
- the following samples from 6/27 were sent to Genewiz for sequencing:
- PyrF F, R (one sample with PyrF_F, one with PyrF_R)
- rpoz F, R (one sample with rpoz_F, one with rpoz_R)
Lambda red results:
- the colonies on the plates did not look promising, and the ones we chose and grew up in LB+kan did not actually grow. Just to be certain, we choose 18 more colonies: 6 from 37.5µL arabinose 100µL plated, 6 from 3.75µL arabinose 100µL plated, and 6 from 37.5µL arabinose 1.5mL plated. Three from each plate were grown in plain LB and three with kan. We will let it grow in 30˚C, overnight if necessary, and hopefully see bacteria for PCR.
- Assuming this does not work, we prepared more ∆HisB∆PyrF∆rpoZ+pKD46 in two ways: we put 3 colonies in LB+amp from the 6/16 transformation plate, and we streaked a new amp plate from the glycerol stock
- Another possibility is that something is wrong with our lambda red. We designed primers to verify that the pKD46 plasmid is really in the cells.
Kan-ZFB-wp-his3-ura3 construct:
- Our last few PCR purifications have given us very low yields, and consequently we have had to use large amounts of our DNA (and the large amounts of buffer salts may also be why our lambda red recombinations have failed). When we tried to amplify our current DNA using the hisura-kan_F and ZFB-wp-hisura_R primers and the Phusion mastermix, it did not work (see 6/23). We will try to gain more product in two ways:
1) Repeat 6/23 PCR but use KAPA mastermix
- the KAPA mix may work better than the Phusion.
- Used KAPA protocol with 1µL of kan-ZFB overlap as template, hisura-kan_F and ZFB-wp-hisura_R primers, 65˚C annealing temp, 90 sec elongation time
- made 2 reactions
2) Repeat overlap extension PCR (see 6/16) with KAPA mastermix
- used KAPA protocol with 1µL kan cassette and 1µL of ZFB-wp-hisura, 65˚C annealing temp, 90 sec elongation
- 10 cycles without primers; hisura-kan_F and ZFB-wp-hisura_R primers added; 15 more cycles
- made 6 reactions
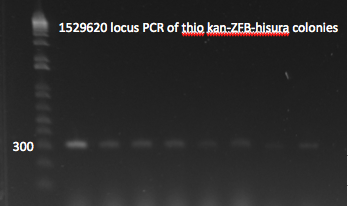
- E gel to check reactions worked: all 6 overlap PCRs successful, but not the other two reactions.
- combined samples 1-3 and 4-6 and ran on 1% agarose gel for extraction
- used Qiagen gel extraction kit and instructions with the following modifications:
- gel bands were dissolved in 500µL of buffer QG regardless of the gel volume
- gel heated at 50C for 20 min (to make up for reduced amount of buffer QG)
- after melting, 10µL of NaOAC (3M) were added to adjust the pH
- DNA from samples 1-3 were eluted in 20µL of ddH2O; DNA from samples 4-6 were eluted in 20µL of buffer EB
- water sample: 273.4 ng/µL, 260/280=1.92
- EB sample: 136.9 ng/µL, 260/280=2.38
June 28th - Bioinformatics
Attention all Harvard iGEM-ers!!! According to the iGEM Main Page, our preliminary project descriptions and safety proposals are due on July 15. Please see the aforementioned link so we can get this done ASAP- we don't want to miss any deadlines and have all our hard work wasted!
- Finalized our Positive Control Sequences, using Justin's macro to insert the F1 helices into the appropriate zif268 F2 backbone
- Length of chip oligos: 131-140bp (based on Cut Site Design)
- Primers: 20bp (x2= 40bp)
- zif268 F2 backbone + helix= 23aa (x3=69bp; some fingers ~3aa longer)
- Some alternate backbones are longer than zif268 F2 backbone
- Type II binding/cut sites= 11bp on each side (22bp total)
- Standard legnth: 40 + 69 + 22 = 131bp
- Use WebLogos as a final visual check of our final generated sequences
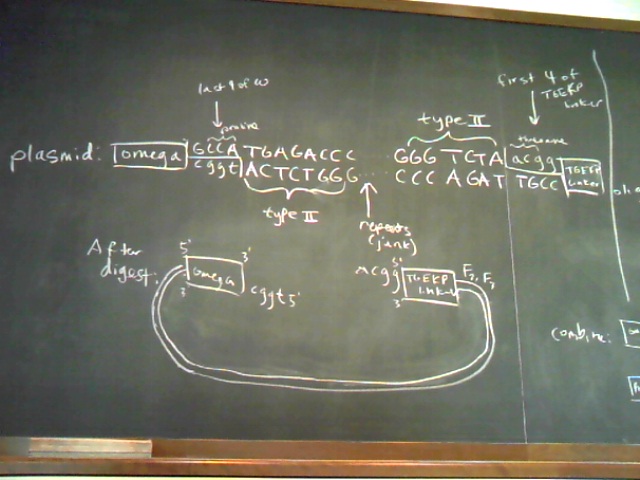
Plasmid and Oligo Design Schematics
Chip-Based Sequence Design Schematic
References
<biblio>
- Kosuri2010 pmid=21113165
</biblio>
Harvard Logo
Running the Generator!
File:HARVFasta total.csv NOTE: LATER GENERATED NEW SEQUENCES. NOT UP TO DATE.
Generated Final Chip Sequences
- We ran the generator once earlier this afternoon, but had to re-run it again due to a typo in the cut sites and the number of sequences we desired for each backbone. Luckily, we caught these errors, and after checking the program once again, we ran it a final time this afternoon.
- It took about 45 minutes for the program to generate and reverse translate the 54900 sequences.
- During this time, we created a function that will re-translate the sequences that the generator output. It compares the original helix with the re-translated helix to make sure that our reverse-translate works properly.
- This step went smoothly, and we verified that the sequences were reverse-translated properly.
- To make sure that the distributions generated were as expected, we made WebLogos of the helices generated(see below).
- The output file (in the Dropbox: iGem > chip > final chip.csv) originally had the following headers: 'Target', 'Backbone #', 'Helix Sequence', 'Backbone Sequence', 'Nucleotide Sequence of Zinc Finger'
- We wanted to convert this information into FASTA format.
- We wrote a function that converted our original file into fasta format (in the Dropbox: iGem > chip > fasta.csv)
- The file FASTA_total (also linked above) contains the FASTA for all 50000 sequences (including the 100 controls).
- For those curious, the FASTA format just a format that looks like this:
- We wanted to convert this information into FASTA format.
>Header (For us the header is: Target, Backbone #, Helix Sequence, Backbone Sequence)(The header for the controls are: Index Number, 'control') Sequence (In our case, the nucleotide sequence of the zinc fingers)
Generated WebLogos for Final Chip
- FASTA-Formatted Chip Data:
- >NNN(Target Triplet) BB# Helix Seq.
- Nucleotide seq. of ZF
Bioinformatics Candids
File:HARVPrimer Index iGEM 2011.xls
Design of Plate Practice Sequences
While we wait for the chip to come in, we have a number of techniques and protocols that we can practice on beforehand, so that when the chip comes we'll be ready to go to use what they give us. We will be practicing the following techniques:
- Cutting ZF1 out of our oligos
- Inserting ZF1 into the expression plasmid in between the omega subunit and the linker before F2
- Verifying that combination of our F1 from the oligo with the plasmid produces a viable, functional ZF
- Amplifying subpools of oligos for testing
- Inserting the expression plasmids into the E. coli containing our selection genome
- Verifying that our ZF-binding site/GFP expression paradigm works
To this end, we will be ordering a 96-well plate from IDT containing oligos that will simulate the entire tube of oligos that we will receive from Agilent in four weeks. These oligos will consist of the following:
- 6 positive controls (we know which DNA sequences these bind to)
- 3 of them being the F1 fingers of Zif268, OZ052, and OZ123
- 3 of them being ZF F1s derived from CODA.
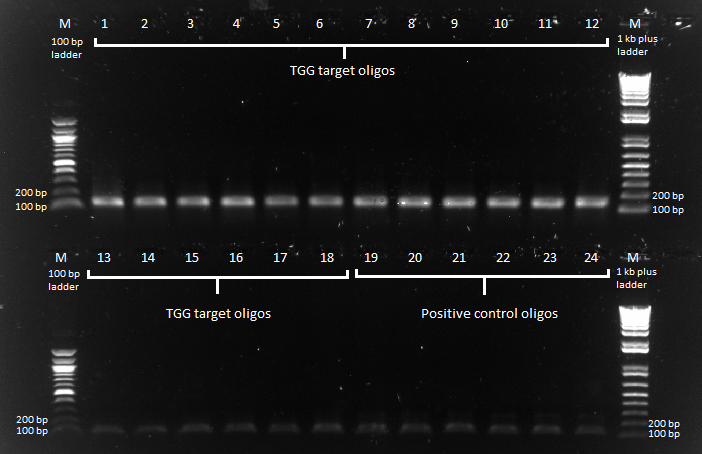
- 90 generated sequences, picked from a subset of the chip
- These are picked evenly across the 9,150 sequences generated on the cihp for the TGG triplet F1 target from the colorblindness "bottom finger" target, GTG GGA TGG. This particular target was chosen because the F2/F1 is a GNNTNN combo, which might be more likely to get hits from our chip generation sequences.
The primer tag sequences for the 90 generated sequence subset will be the same as they are on the chip (for the sake of explanation, we will refer to them now as P1F and P1R in this paragraph). The positive controls will be flanked immediately by the same primers as the generated subset so that we can amplify everything as one pool altogether should we need to (so this will be P1F and P1R). However, we will also put an additional set of primers outside of the P1F/P1R primers for the positive controls so that we can specifically amplify the positive control subpool, should we want to. These primers will be the same as the primers for the positive control on the chip (which will be called P2F and P2R here).
To recap, on the chip we will have the following oligos :
45750 other oligos for the 5 other target sequences Oligos (TGG set, 9150 total): | P1F | type II binding site | generated F1 | type II binding site | P1R | Oligos (+ control, 100 total): | P2F | type II binding site | control F1 | type II binding site | P2R |
In our test pool of 96 sequences, we will have two types of oligos (note the two pairs of primers around the positive controls):
Oligo (TGG set, 90 total): | P1F | type II binding site | generated F1 | type II binding site | P1R | Oligo (+ control, 6 total): | P2F | P1F | type II binding site | generated F1 | type II binding site | P1R | P2R |
Once we get our test sequences back from IDT, they will come in a 96-well plate with one oligo in each plate. We should make a mixture using some of each well in order to create a tube that contains all 96 sequences. This will simulate the tube that we will receive from Agilent, except instead of 55,000 sequences we will have 96 sequences only in this tube. From here, we can practice using this as a library.
We can pretend that this tube is just 96 generated sequences on the chip, treating the positive controls as if they were also generated sequences (we only include them in the 96 to ensure that we will indeed get a "hit" from this practice screening). Thus, we can just use the P1F/P1R primer set to amplify all of them in order to use them for the subsequent steps.
These subsequent steps will be those that were outlined above, namely cutting out the F1 sequence from each oligo, ligating this F1 into our expression plasmid, putting the expression plasmid into our selection strain, observing colonies which get infused with ZFs that bind to our target site (the "hits"), and sequencing the colonies that get hits to determine which ZF they are expressing.
We will be repeating these exact same steps once we get the chip, so if we can perfect our protocols with these practice sequences, we should be golden when the chip comes in.June 29th
Our first day with everyone in the wet lab!
PyrF and rpoZ sequencing:
- For some reason, Genewiz said that sequencing failed due to "no priming." We will redo the PCR and send the products in again today.
Lambda red recombination and MAGE:
- The cultures made from the kan plates from our earlier attempt at lambda red did (in some cases) grow, including one colony in kan from 3.75µL arabinose, 100µL plated.
- PCR of liquid culture: used 1µL of culture diluted 1:20 as well as saturated culture of ∆hisB∆pyrF∆rpoZ+pKD46 (diluted 1:20) as a 1529620 locus wild-type control
- primers (from Vatsan): 1529481-f, 1529806-r
- KAPA mastermix and procedure with 56˚C annealing and 90 seconds elongation
- E Gel of product: both wild-type and the sample hopefully containing the insert had the same short band of around 350bp--the recombination was unsuccessful
- PCR of liquid culture: used 1µL of culture diluted 1:20 as well as saturated culture of ∆hisB∆pyrF∆rpoZ+pKD46 (diluted 1:20) as a 1529620 locus wild-type control
- Used overnight saturated culture to reinoculate; once close to mid-log, culture split into 2 1.5mL amounts and 37.5µL arabinose added to each
- same procedure as previously described, but with about 300ng kan-ZFB construct and a 2.5µM final concentration of HisBNuke3 (12.5µL of 10µM stock)
- electroporate with 1.8 kV, about 5
- recover 3 hrs
- kan-ZFB insertion colonies plated on kan, MAGE on amp
PCR Preparation
- Lambda Red- Selection strain glycerol stock: 1/100 dilution, 2 uL stock with 198 uL ddH2O
- Spec (Colony)- Touch 1 colony with pipet tip, add and mix with pipet in 20 uL ddH2O, then vortex
PCRs
- PKD46 (Lambda Red)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 1 uL
- ddH2O- 10 uL
- (25 uL total)
- Spec (Colony)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 1 uL
- ddH2O- 10 uL
- (25 uL total)
- Spec (Miniprep)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 2 uL
- ddH2O- 9 uL
- (25 uL total)
Expression Plasmid Design in silico
Today, we designed our expression plasmids in SeqBuilder. This included plasmids for our 6 target sequences, and 3 positive controls (9 in total). These positive controls were the following:
| Index | Nucleotide Sequence (5\'-3\') | Helices (F3 to F1) | Notes |
| 16 | GAA GGG AAC | QDGNLGR RREHLVR HRTNLIA | Very similar to one of our target sequences (CB top), which is GAA GGG ACC |
| 55 | GGA GTG GTG | QTTHLSR DHSSLKR RNFILQR | Very similar to a target sequence (FH top), which is GGA GTG CTG |
| 77 | TGT GAA TAG | RRRNLQI QQTNLTR QPHGLTA | Out of ze air |
Each of our expression plasmids contained:
- Omega subunit
- Omega/F1 linker (taken from paper that Dan and Noah emailed us), http://nar.oxfordjournals.org/content/36/8/2547.short
- type II binding sites
- gap between type II binding sites that contains XbaI restriction enzyme site (which is not present anywhere else in the entire expression plasmid)
- F1/F2 TGEKP linker
- F2 for a specific target sequence
- F2/F3 TGEKP linker
- F3 for a specific target sequence
- TAA stop codon immediately after F3
All of this is on our spec-resistance-containing plasmid. The above construct replaced the GFP which was present previously on this plasmid.
Tomorrow we will begin design of our primers from these SeqBuilder files.June 30th
Lambda Red, Backbone, and Sequencing PCR
- Gel run on the presence of a Lambda Red protein in the pKD46 plasmid showed that it is indeed present, so our recombination failures have not been due to an incorrect plasmid.
- Gel run on the backbone of pZE21G plasmid was success and took us one step closer to obtaining all parts necessary for the three part assembly
- Gel run on the pyrF and rpoZ was success
- Therefore we sent the PCR products and primers to GENEWIZ for sequencing again
pZE21G backbone:
- Since last night's PCR was successful, we will redo it with a few protocol adaptations to get a cleaner product and to increase our yield when we purify
- KAPA mastermix and protocol: primers HindIII-F and KpnI-R
- template: pZE21G miniprepped plasmid, 1µL
- 2 min elongation time, 30 cycles
- 2 samples at 55˚C annealing, 2 samples at 60˚C
Lambda red and MAGE:
- Yesterday's prep produced tiny colonies on the MAGE plates and (so far) none on the kan-ZFB plates. Just in case it didn't work, we will redo the lambda red using even more DNA and perform a second round of MAGE using culture from yesterday that was not plated.
- same procedure, but with the following changes:
- 5µL kan-ZFB (about 1 mg)
- recover 3 hrs
- kan-ZFB: plate 100µL and 2 mL on kan plates
- MAGE: plate 1µL and 10µL on amp plates
- To see if the colonies on the MAGE plate knocked out HisB, we chose 24 colonies, resuspended them in water, and put half the cells in LB (complete media) and half in NM media (does not have histidine). 96 well plate, 150µL media, grown overnight at 30˚C.
ZF Expression Plasmid Ultramer and Primer Design
Today, we designed primers ZF_073 through ZF_085 as listed in the iGEM Primer Index spreadsheet. These were basically two sets of primers: the primers to clone out the omega subunit and linker, and the ultramers that would construct the last part of the linker along with the type II binding sites and F2/F3 fingers. One should refer to the primer list for the sequences.
Note: the annealing sequence for the ultramer overlap contained a 72 degree melting temp hairpin. To get around this, I changed one of the codons in the F2 backbone. The F2 backbone begins with "FQCRIC", and so I changed the codon for the arginine (R) from CGC to CGT, which resolved the hairpin problem.
June 30th - Bioinformatics
Updated Primer list and FASTA formatting
We ran into a small hiccup, when we were informed that we had forgotten to reverse translate the reverse primer sequences that were being appended to the generated sequence. This is because the primer sequences we were given were the sequences for the actual primers, rather than sequences to which the primers would bind. Luckily, we caught this error! We did have the re-run the generator because we had to make sure that our generated sequences did not contain the new primers.
- Here is the updated primer list:
This is the set of final target sequences with assigned forward and reverse primers (tags for PCR):
| Disease | Target Sequence | Forward Primer (5'-3' NOT REVERSE COMPLEMENT) | Reverse Primer (5'-3' REVERSE COMPLEMENT) |
| Colorblindness | GCT GGC TGG | ATATAGATGCCGTCCTAGCG | TGGGCACAGGAAAGATACTT |
| Colorblindness | GCG GTA ACC | CCCTTTAATCAGATGCGTCG | GGTCGCCCTTATTACTACCA |
| Familial Hypercholesterolemia | GGC TGA GAC | TTGGTCATGTGCTTTTCGTT | TCTGAGTATCCGATACCCCT |
| Familial Hypercholesterolemia | GGA GTC CTG | GGGTGGGTAAATGGTAATGC | GCTATATCCGGGGAATCGAT |
| Myc-gene Cancer | GGC TGA CTC | TCCGACGGGGAGTATATACT | TTGGCCTGAAGCAGTTAGTA |
| Myc-gene Cancer | GGC TGG AAA | CATGTTTAGGAACGCTACCG | GGGAGGGAACGGAGATTATT |
| Controls | n/a | GTACATGAAACGATGGACGG | CGCTGAGGAGACTATACCAG |
There was also a small error in the FASTA formatting. There are not supposed to be any spaces in the header, so the spaces were replaced with underscores.
- Example:
>1_controlGTACATGAAACGATGGACGGGGTCTCAGCCATTCCAATGTCGTATCTGTATGCGTAATTTTTCACGCAAACACCATTTGGGTCGTCATATCCGTACGCACACGGTGAGACCCGCTGAGGAGACTATACCAG
July 1st
HisBNuke3 MAGE results:
- All the MAGE round 1 colonies grew overnight in the LB but not the NM medium: either they all had the HisB gene knocked out (unlikely) or there is something wrong with the NM medium. As a control we grew (∆HisB)∆pyrF∆rpoZ+pKD46 in NM, but nothing grew, implying that the problem is with the NM medium.
- There were small colonies on the MAGE round 2 amp plates, so we chose 12 from each plate and grew them in 100µL of SOC medium.
- PCR of the colonies for sequencing:
- KAPA mastermix and protocols, with His3_F and His3_internalR primers, 2µL culture as template, 65˚C annealing, 15 sec elongation, 25 cycles
- ran on gel: band around 500-600, but very faint (probably because we did not let the culture grow up enough)
- sent to Genewiz to be sequenced with His3_F primer
Lambda Red recombination results:
- The plates from 6/30 had an almost crystalline formation on them, which may or may not be bacteria. The 6/29 plates, which we had left at 30˚C, did have colonies but they may be contaminants. We grew up 12 colonies from each 6/29 plate and 6 "colonies" from the 2mL 6/30 plate in SOC medium with kanamycin, and next week we will PCR the 1529620 locus to check.
pZE21G spec backbone:
- PCR of backbone was successful (see E gel below). Gel extracted with Qiagen kit with the following modifications to the kit instructions:
- 2 gel volumes of buffer QG, heated 10-15 min
- after heating added 10µL 3M NaOAc
- eluted in 30µL ddH2O
- 157.4ng/µL 260/280=1.9; 129.2ng/µL 260/280=1.88
Selection system plates:
- Made 3-AT and 5-FOA plates following 2006 Meng Nature Protocol (see protocols for details). 5-FOA plates have a blue stripe, 3-AT plates (0.5 mL 3-AT) have a green stripe.
July 4th
FIREWORKSJuly 5th
Team ZF Assembly
- Assemble and confirm zif268 plasmid
- Lac-ω-zif268 with spec → Lac-zif-ω with cpic
- ZF: 2-3 ultramers
- PCR out ω subunit
Team Wolfe Selection
Goals for the week:
- MAGE out HisB
- What we've been trying/why it's not working — keep trying!
- PCR out Kan Cassette (colony PCR)- analysis/check that it worked
- We did the insertion and have things growing in Kan that look like our cells — we probably have it!
- As long as it's the right size and in the right place, it probably worked, meaning we have 2 HIS genes → MAGE is to remove the endogenous genome copy in vivo
- Construct (PSR01): ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~
- Fix/Test NM media
- Made last week, nothing worked in it — try more glucose
- Wolfe strain seems to grow really slowly (~36h); try glycerol stock of PKD46, grows faster
- Make other 3-AT plates (1.5)
- 3-AT is a competitive inhibitor of His; the stronger His is expressed, the more 3-AT the cell can tolerate and still grow
- Test 3-AT plates
Today's results:
- PCR to check whether kan-ZFB-wp-hisura was successfully inserted into the genome
- Used KAPA master mix and protocols, same procedure as 6/29/11
- Because of KAPA shortage, only ran samples 6-8 from 100µL 6/29 plate and 6-7 from 2mL 6/29 plate plus (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture as a control (should produce a band around 300bp, while the insert would make the band 2-3kb)
- Ran on E-Gel with team TolC's samples, but there was an issue loading the gel, so also ran separately on another gel. Unfortunately most of the PCR reactions strangely did not work, but two of the samples appeared not to have the insert (see below).
- Redo PCR:
- out of KAPA so had to use Invitrogen Platinum Supermix
- each reaction had 22.5µL supermix, 0.5 µL each 10µM primer, 1µL template, 0.5µL H2O
- 20 reactions, including one from the same control used above.
- followed Invitrogen instructions, with 3 min elongation, 56 C annealing, 30 cycles
- Assuming the PCR is unsuccessful, we will grow up (ΔHisB)ΔpyrFΔrpoZ+pKD46 (from 6/28 plate) overnight in LB+amp
- NM medium:
- To test Sarah's NM (made 6/30) we will grow up pKD46 strain in 3mL of it overnight at 30 C. If no bacteria grew, we will try remaking it ourselves.
Team TolC
- PCR out and insert Kan construct in front of TolC (similar to Wolfe selection)
- Construct 1: ~ - KAN - ZFbs - weak promoter - ~ (done, PCR, gel extraction)
- Used primers ZFB-wp-tolC-r and tolC-kan-f
- Construct 2: ~ TolC ~ (do not have primers yet)
- Note that ~ indicates a homology region
- Construct 1: ~ - KAN - ZFbs - weak promoter - ~ (done, PCR, gel extraction)
- Need the EcNR2 strain to come in before we can work with it (came in Thursday 7/7/2011, the one from Wednesday 7/6/2011 was not growing well.)
- Inactivate rpoZ (similar to inactivating His3, but should be easier to use MAGE with- EcNR2 is a strain designed for MAGE)
- MAGE oligo rpoZ deletion will arrive on Thursday 7/7/2011
- Make SDS (how we test TolC +vely)(done, stored at room temperature)
- Only cells with active TolC can survive SDS treatment
Web Design
To Do
- Safety Description
- Email Alain/Harvard biosafety group ✔ (Meeting pending)
- Needed information:
- Strain names
- Antibiotic Resistance
- DNA inserts/removals
- Draft Project Description ✔
- Finalize
- Update the Facebook page/create a new 2011 page
- Get the Public Wiki up and running
- Figure out templates
- Update Twitter
Notes to All Teams
- Teams: Make sure everything is organized in your dropbox!
- *Announcing a New (and Final) Plasmid Name* PSR01: formerly known as Wolfe Plasmid/Vatsan's Plasmid/Selection
- Contains ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~
July 6th
Team ZF Assembly
Because our primers still haven't arrived, the members of Team ZF Assembly are working with the Web Design Team.
Team Wolfe Selection
- Redo PCR Results:
- Samples 1,3,4,5,6,7,8,9,11 from the 100µL 6/29 plate all showed unsuccessful introduction of kan-ZFB-wp-hisura
- Samples 2,3,4,6,7,8,9,10,11,12 from the 2mL 6/29 plate all showed unsuccessful introduction of kan-ZFB-wp-hisura
- (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture used as a control
- NM media has 1% of the glucose concentration written in the protocol, it is 0.4mg/ml instead of 40mg/ml. Nevertheless, our overnight culture of pKD46 grew successfully so we will continue to use the media unless other problems arise.
- One possible explanation for our failure to PCR out a kan-ZFB insert from the recombined colonies is that the insert was integrated into the genome somewhere else besides the 1529620 locus. This might be why the PCR shows no insert while the colony still grows in kanamycin. We will try to PCR the insert using primers that anneal to the kan and his-ura sections (since these primers are about 70 bases long, we will do a 2-step cycle)
- Invitrogen Platinum Supermix reagents and protocols, with 3:30 at 72˚C instead of separate annealing and elongation, 25 cycles
- used same cultures as for PCR listed above
- kan-ZFB-wp-hisura insert as control
- results from 2 E Gels: PCR not very successful. Colonies showed no bands while the control showed only a side product too small to be the full insert.
- Grew up 24 more kan-ZFB lambda red recombined colonies (12 from 100µL 6/29 plate, 12 from 2mL 6/29 plate) in LB+kan in 96 well plate for PCR tomorrow
- Diluted 24 colonies (1 ss+pKD46 control, 11 from MAGE round 2 1µL plate, 12 from MAGE round 2 10µL plate) in 30µL ddH2O and split between NM,NN+Histidine, and LB+amp to check his phenotype
- NM+his media: 49mL NM, 1 mL 5% histidine solution
- Made 1.5mL 3-AT plates
Team TolC
- Gel extraction of KAN-ZFBs-wp as there is a side product.
- To increase yield, spin down the contents of both eppendorf tubes into a single column, have a 2:1 volume ratio of QG Buffer to gel volume, e.g. 1ml QG buffer to 0.5ml gel in step 2, and elute in 30µl water in step 13)
- checked the ECNR2 strains, both appear to have not grown
- note from 7/7/2011, new ECNR2
- running another PCR since there was errors in gel extraction, and the yield was less than 20ng/µl, and purity was very bad.
TOLCINV PCR-Machine 7
- 94°C for 2 min
- 94°C for 30 s
- 53°C for 30 s
- 72°C for 1.5 min
- Previous three steps repeat 30 times
- 72°C for 5 min
- 4°C forever
Web Design
- Project description
- Web design brainstorming;exploring past designs and templates
July 7th
Team ZF Assembly
Our primers still haven't arrived. We are working with the Web Design Team.
Team Wolfe Selection
- PCR for Round 2 MAGE cultures in LB media
- The samples came from the overnight culture of MAGE/control strains grown in LB+amp, NM+his, and NM. Oddly, the (∆HisB)∆pyrF∆rpoZ+pKD46 control grew only in NM+His and LB, and not in plain NM.
- Samples
- Control= 1 ss+pKD46
- 11 cultures from 1μL plate in LB media
- 12 cultures from 10μL plate in LB media
- Program
- Anneal at 56°
- Elongation for 90 seconds
- All else according to standard KAPA protocol
- E gel of control and 1µL plate colonies 1-8 to check PCR worked (it was successful)
- samples prepared for sequencing tomorrow
- Kan-ZFB insert check PCR results:
- the overnight cultures grown yesterday were used in a PCR (same KAPA protocol as before) around the 1529620 locus. Not all the cultures grew; 100µL plate cultures 1-5 and 2mL plate cultures 1-12 were used.
- Results: some of the reactions, strangely, did not work, but the ones that did showed a short band and thus did not have the insert (see below)
- HisBNuke3 MAGE round 3:
- used same protocol as previously described, using 1.5mL mid-log culture from MAGE round 2
- will recover overnight so that we can reinoculate and continue MAGE rounds tomorrow
- NM medium:
- added 3.9g glucose to 100mL NM medium to bring the glucose concentration up to the correct level.
- Because (∆HisB)∆pyrF∆rpoZ+pKD46 did not grow in plain NM, we are curious to see if that was due to low glucose levels. One colony from a plate was put in 3mL NM(gluc) and will be grown overnight.
- HisB MAGE2:
- In case the sequencing does not show successful MAGE, we will grow overnight 48 more colonies (24 from each plate) in 100µL of NM(gluc) and LB+amp
Team TolC
- making culture tubes of ECNR2 and ECNR2delTolC for glycerol stock (no Amp)
- making culture tubes of ECNR2 for MAGE to delete rpoZ (one with amp, one without)
- dilutions of rpoZ deletion MAGE oligo, 20µM
- dilution of rpoZ deletion MAGE oligo, 1µM, for MAGE.
- serial dilution of ECNR2 after MAGE round 1, and round 2, with 20µl of 100x dilution of concentrated culture. For plating onto amp plates.
- During Gel extraction, ignored 3x volume in step 2 and instead used 500 µl of QG buffer. Additionally, used 18 µl of EB buffer and not water in step 13
Web Design
Brainstorming
- Minimalist toolbar, metallic crimson color
- "Popout" menu below, not dropdown
- Slideshow on front page
- First slide: Brief text project description, brief youtube video description (in layman's terms)
July 8th
Team ZF Assembly
PCR of omega+zif268 with overhangs
We performed a PCR to take out our omega subunit plus zif268. We used the following recipe:
- 10 ul KAPA readymix
- 0.75 ul of hindIII (primer)
- 0.75 ul of wF+plLacO (primer)
- 1.2 ul template
- 12.3 ul water
The first steps of the PCR involved heating at 95* (for 5 minutes) and 98* (for 20 seconds). Annealing temperature was 56*. Extension time was 30 seconds.
The product of the PCR should be 709 bps. It includes the omega subunit, zif268, as well as overlap sequences homologous to the selection plasmid. We ran an e-gel to ensure that our desired product was present:
Gel purification of PCR product
We had to gel purify our PCR product in preparation for isothermal assembly, using our protocol for gel purification. The result from running the gel can be seen below - we cut out the bands at 709 bp, the expected size of our product.

In the final step of the purification process, we eluted our DNA in buffer EB. We used the nanodrop spectophotometer to determine the concentration of DNA present.
- The concentration was 54.3 ng/ul.
Team Wolfe Selection
MAGE round 3:
- MAGE cultures that recovered overnight were saturated, so we made 1:1000, 1:10000, and 1:100000 dilutions and plated 20µL of each onto amp plates and left at 30˚C overnight.
MAGE round 2 cultures from yesterday:
- We did a PCR (His_F and and His_R primers) on 24 of the 48 cultures we grew and sent them off for sequencing.
Results of NM culture:
- The selection strain (∆HisB∆pyrF∆rpoZ+pKD46) did not grow in NM, which is worrisome. None of the MAGE samples grew in NM either, so either they all have HisB knocked out or there is something wrong already with the strain's His pathway.
- We will grow up selection strain (without pKD46) from glycerol stock in LB+tet and transform with Vatsan's plasmid tomorrow to see if that has any effect on the his-deficiency phenotype.
Kan-ZFB insertion: modified nucleotides:
- Primers finally arrived that would put modified nucleotides on the 5' ends of the insert, which should improve lambda red efficiency.
- PCR: KAPA protocol with Kan forward phosphorthioate and URA2 reverse phosphorthioate, 56˚C annealing, 90 sec elongation, 25 cycles, 1µL kan-ZFB-hisura DNA as template
- Results: PCR only produced a smaller side product. This is strange since we also ran a gel of all our different kan-ZFB-hisura purifications and they all run at the correct size (see images below).
Team TolC
- Performed re-inoculation of ECNR2-Placed into tube labeled ECNR 2 MAGE 3 in 30 degree shaker.
- grew up 24 colonies total for sequencing ECNR2 for rpoZ deletion (the colonies appear to have grown much faster than expected).
- rows A (MAGE round 1) and B (MAGE round 2), for PCR and sequencing. to be placed in the 4C refrigerator after use as template.
- RPOZ PCR-Machine 6
- Primer Annealing = 56 degrees
- Extension 48 = seconds
Team Web Design
- Edited/Re-drafted our project description, see Dropbox (It's a bit long as is now, but the excess informaiton that we cut out can still probably be used somewhere on our website
- Sketched draft pages of the public wiki
- Homepage- first draft sketch done based on the characteristics and requirements we'd discussed. However, there are a few issues (i.e., how to include some of the iGEM wiki reuqirements).
July 9
Team ZF
Isothermal Assembly
We performed an isothermal assembly in order to insert omega+zif268 into the plasmid containing the spec backbone.
- The concentration of the plasmid (previously prepared) was 157.4 ng/ul.
- The concentration of the w+zif268 was 54.3 ng/ul.
Using our isothermal assembly protocol we determined we needed:
- 0.63 ul backbone (length: ~2.2 kb)
- 0.59 ul w+zif268 (length: ~0.7 kb)
- 2.78 ul water
- 15.0 ul isothermal assembly solution
Transformation of electro-competent E. coli
We electroporated our cells so they would take up the plasmids, using:
- 1.0 ul DNA
- 40.0 ul cells
- 500 ul LB
We plated the cells, using this protocol, on spec plates, one of which contained 50 ul of cells and the other 150 ul of cells. The cells have been incubated at 37*, and will be left there overnight. Cells that have taken up the plasmid have spec-resistance and should be able to survive. Tomorrow, we will be picking colonies and performing PCR on them to determine whether they contain w+zif268.
Team Wolfe
- MAGE3 plates look good--we'll choose colonies for sequencing on Monday.
- Transformation of selection strain with Vatsan's plasmid:
- We're curious to see whether Vatsan's plasmid (Zif268 omega, ZFB, wp, hisura) will have an effect on the (∆HisB)∆pyrF∆rpoZ strain's growth despite the seemingly intact HisB.
- Transformed mid-log selection strain (reinoculated from overnight culture) with about 100ng of Vatsan's plasmid following standard procedure.
- After 2 hr recovery plated on amp plates with 100µL or 1mL
July 10th
Team ZF
PCR to check for omega+zif268
All colonies that grew on the plates we left overnight should have spec-resistance, but they may not have the omega and zif268 unit that we inserted yesterday with isothermal assembly. We performed a PCR on 13 colonies picked from the 50 ul plate to find a colony that contains the w+zif268 insertion. We used the same recipe at our PCR reaction [[#PCR of omega+zif268 with overhangs on Friday. If the insert is present, then we should see a band at 709 bp. We also included 2 positive controls: our isothermal assembly product and our selection strain from which the w+zif268 was originally copied.
We ran an agarose gel to check the bands:
July 11th
Team ZF
We did a miniprep of the 12 colonies we picked Sunday and left overnight in LB. Here are the concentrations of our products:
| ID | Conc. (ng/ul) | 260/280 |
|---|---|---|
| 1 | 6.63 | 1.27 |
| 2 | 12.96 | 1.53 |
| 3 | 9.28 | 1.41 |
| 4 | 7.23 | 1.31 |
| 5 | 12.64 | 1.55 |
| 6 | 10.77 | 1.48 |
| 7 | 17.19 | 1.73 |
| 8 | 39.70 | 1.78 |
| 9 | 55.73 | 1.81 |
| 10 | 24.34 | 1.72 |
| 11 | 25.19 | 1.68 |
| 12 | 38.57 | 1.74 |
| 13 | 27.26 | 1.75 |
Then we performed a PCR on the 12 miniprep products, the 12 colonies in LB, as well as 3 controls (negative control, isothermal assembly product, selection strain) to find a colony that has the omega+zif26 insertion:
- 10 ul KAPA readymix
- 0.75 ul of hindIII (primer)
- 0.75 ul of wF+plLacO (primer)
- 1.2 ul template
- 12.3 ul water
The first steps of the PCR involved heating at 95* (for 5 minutes) and 98* (for 20 seconds). Annealing temperature was 56*. Extension time was 30 seconds.
We ran a 1% 100 ml. agarose gel at 120V to check the success of the PCR.
The bands were rather thick, so it was difficult to estimate the exact band size. However, the bands appeared a bit below 650 bp, although we expected a size of 709 bp, so we ran an e-gel on some of the colonies to ensure that that bands were indeed the right size.
The bands were closer to the size that we expected, so we decided that we successfully inserted w+zif268 into the plasmid. We decided to pick colony 9 (which had the highest concentration post-miniprep) and work with that from now on. We made spec plates with 50 ul of colony 9.
Team Wolfe
Selection strain transformed with pSR01:
- both plates had lots of colonies--almost too many. We need to make sure the plasmid is, indeed, in these cells.
- Colony PCR:
- picked 6 colonies and diluted in 7µL H2O: 3 for PCR template and 4 to be grown up in LB+amp
- KAPA protocol using M13_F and M13_R primers (anneal to the vector), 50˚C annealing, 2min elongation
- E gel results: lots of bands but rather messy and not exactly what we'd expect. We'll try to verify the presence of the plasmid in a few other ways.
- Sequencing:
- We sent out the PCR samples with M13_R to Genewiz
- Culture PCR:
- We used the cultures made with the colonies used for PCR above as template to see if this gives us cleaner results.
- same protocol as above except with 1µL of 1:20 dilution of culture as template.
- Assuming that the plasmid is indeed in the colonies, we will grow an overnight culture in LB+amp as well as the Wolfe selection strain and tomorrow we will use a growth reader to check their phenotypes in NM
MAGE round 3 colonies:
- In case pSR01 does not rescue the selection strain's growth phenotype, we will PCR 96 colonies from the MAGE3 plates and send them out for sequencing tomorrow.
- Used 9µL KAPA, His_F and His_R primers, 3µL suspended colony (out of 7--the remaining 4 grown in LB+amp)
Thio kanZFB:
- We retried the PCR to put modified nucleotides on the ends of the kan-ZFB-wp-hisura insert using the same procedure as 7/8/11
- E gel: reactions didn't work
- Tried again with a longer elongation time (3 min) and let run overnight.
Team TolC
Sequencing of rpoZ delete
- Since the sequencing was sent after 3pm on Friday, it wasn't picked up before today (Monday), so we are going ahead with lambda red recombineering of the KAN-ZFB-wp into the ECNR2 genome
- glycerol stocks of ECNR2 and ECNR2ΔTolC were made.
Team Web
- Looked at good wikis from last year.
July 12th
Team ZF
PCRs of finger ultramers
We received the ultramers for fingers two and three of our 6 experimental expression plasmids. We performed an overlap PCR in order to join the two ultramer pieces and make a whole unit. Ultimately this unit with be assembled with the omega subunit and the spec backbone to make our final expression plasmids. We used the following recipe:
- 10 ul KAPA
- 13.5 ul water
- 0.75 ul forward ultramer
- 0.75 ul reverse ultramer
We had an annealing temperature of 58* and an extension time of 15 seconds.
However, in addition to the expected band at approximately 300 bp, the bands for our ultramers was extremely streaky, indicating that there may be other contaminants. We decided to perform a PCR purification of these samples, using our protocols, and see if there was an improvement in the gel.
Even after the gel purification, our bands were streaky. We decided to redo the PCR, placing all the reactions in single tubes, performing a touchdown PCR instead. Our annealing temperature started at 70*, repeated for 2 cycles, and then decreased 2*, every two cycles, until it reached 58* (the optimal annealing temperature), where it remained until the 25 cycles were complete. We left this PCR going overnight. (See July 13th for the gel image.)
PCR of expression plasmid cross-junction
We also performed a second PCR on colony nine (the colony we decided with which we decided to proceed) to confirm that our omega+zif268 in the spec backbone was successfully inserted into the E. coli. The expected product was the cross-junction on the plasmid, approximately 1.4 kb. We used the same recipe as that used on July 9th, except with a forward primer of PZE23G-3581 F and a reverse primer of PZE23G-2133 R, and had a 55* annealing temperature and an extension time of 45 seconds.
After running an e-gel, we confirmed that our w+zif268 was inserted into the plasmid.
PCR and gel extraction of omega subunit
We performed a PCR to get out the omega subunit+linker, with an annealing temperature of 56* and an extension time of 15 seconds. The miniprep from colony 8 was the template DNA. We will use the product in assembling our final expression plasmids.
We determined that the PCR was successful after running the sample on an agarose gel, and performed a gel extraction so that we could use the product in our isothermal assembly.
The gel extraction was successful, and we had a final concentration of 120.5 ng/ul.
Overlap PCR of OZ052 and OZ123
We performed an overlap PCR using the ultramer pairs that solely encoded the F1, F2, and F3 fingers of OZ052 and OZ123. The annealing temperature was 62* and the extension time was 15 seconds. The results of the PCR can be found below (the products are only in the last two lanes) and the products were also PCR purified before being put into storage at -20*C.
Team Wolfe
MAGE round 3 samples:
- 94 sent out for sequencing (numbers go A1-H1, etc.)
Thio kan-ZFB results:
- PCR run out on E Gel: not successful. We do not know what is still causing our PCR to fail.
pSR01-transformed selection strain:
- We discovered that hisB is a two-function protein, with the N-terminal region acting as an HPase and the C-terminal region as an IDPG. His3 only complements the IDPG activity, so even if our MAGE worked, it might destroy our selection system. The two aa deleted in hisB are in the C terminal region, so maybe they do inactivate it. We emailed Keith Joung whose lab made the selection strain to see how HisB was deleted.
- In case this mutation is correct but lambda red in the selection strain still eludes us, we designed MAGE oligos for use in EcNR2 to either delete the two aa or put 3 stop codons after the HPase region.
- culture PCR results: cultures 3-4 had a strong band (and 2 and 5 weak ones) around 850-1000bp. It is interesting how different these results are from yesterday's colony PCR gel.
- Sequencing results: The sequences sent from Genewiz aligned well with the pSR01 plasmid sequence and covered pretty much all of URA3. This implies that the cells did indeed take up our plasmid.
- Growth phenotype:
- 12 colonies were chosen and split between NM (gluc), NM+IPTG (to induce zif268 expression), NM+his (gluc), NM+his+IPTG, LB+amp, LB+amp+IPTG and grown overnight. (Final concentration IPTG=500µM)
Team TolC
- MAGE knockout sequencing
- Received sequencing from the MAGE knockout of rpoZ within the TolC genome
- Observed all 24 sequenced attempts and it appears as none of them have the knockout and stop codons placed into genome
- Possible reason behind failure is that one, we may not have been reading the results correctly or two, the MAGE oligo had all three stop codons right next to each other in the oligo and may have needed to be separated by three nucleotides each in order to maximal annealing
- Lambda Red Recombination of kan-ZFB-wp insertion into MAGE knockouts
- Plates from the recombination showed small colonies
- With this we picked 6 colonies from the 4 different plated recombination cultures and grew them to saturation
- Began PCR with TolC_Seq_F and TolC_Seq_R primers in order to determine whether recombination was successful
- Procedure (on gradient PCR machine, name: ABC, folder: igem)
- KAPA mixture
- Then 95 C, 5 min
- 98˚ C, 20 sec
- 60˚ C, 15 sec
- 75˚ C, 45 sec
- Repeated cycle for 30 times
- 72˚ C, 5 min
- 4˚ C, forever
- Tomorrow we plan to run gel
- Length will be:
- 450 bp if insertion failed
- 1200 bp if insertion of Kan cassette only
- 1650 bp if insertion of Kan-ZFB-wp
- Length will be:
- Plates from the recombination showed small colonies
- Insertion Construct for TolC (homology-kan-ZFB-wp-homolgy)
- Began 90 µL total of PCR for the insertion construct since we ran out
- If gels show failed PCR tomorrow we will start from normal ENR2 without any MAGE and begin with lambda red recombination and possibly make new MAGE primer and then knockout rpoZ
Team Web Design
- Met with Harvard biosafety officer, Sidney
- Send to him: list of strains, plasmids that we are using
- Note to all iGEM-ers: We will all be attending a biosafety seminar on Friday, July 15th from 10am to 12n in Biolabs 1058.
- In case you haven't already, please see Alain's message below:
- Every member of the Harvard iGEM team (unless you are absent on Friday) has to attend the next lab Safey/Biosafety Blood born pathogens training this coming Friday 7/15/2011 from 10 am to 12 noon. The training takes place in the Biological laboratories (16 Divinity Ave) room 1058. If you cannot go and have a very good reason let me (Alain) know. If you attended a similar training session on the Cambridge campus or at the Medical school you don't have to attend. Otherwise you have to go.
July 13th
Team ZF
Results of ultramer touchdown PCR
We ran an e-gel on the product of our touchdown PCR. Once again we have our band at 344 bp. It is still slighly streaky, but much less than our earlier PCR reaction. There are some faint bands smaller than our expected product, which are likely the ultramers we used in the reaction. We'll use the product for our isothermal assembly later today.
Isothermal assembly and transformation of expression plasmids
We followed our protocol for isothermal assembly, using the following recipe:
- 0.63 ul backbone (at 157.4 ng/ul to get 100 ng)
- 0.13 ul omega+linker (at 120.5 ng/ul)
- 0.50 ul ultramer (~30 ng/ul)
- 1.24 ul water
- 7.5 ul isothermal assembly solution
The sizes of the pieces we put together were the following:
- Backbone: 2.2 kb
- omega+linker: 352 bp
- ultramer: 344 bp
Note: We also made a negative control of the isothermal assembly, only putting in the spec backbone without omega+linker or ultramer.
We then performed transformations to insert our isothermal assembly products into E. coli, following our usual protocol. However, we ran out of electrocompetent cells, so only samples 78 - 82 were done with electroporation while 77, 83-85, and the negative control were done with chemical transformation, using these protocols (the samples are named after the reverse ultramer ID in the primer index that were used to generate the ultramer sequences).
We then plated 1 ul of the cells on spec plates and left them overnight at 37*. Because we ran out of our own spec plates, we also had to use LB plates that had spec added to it, and spec plates that were made in April.
Team Wolfe
∆HisB∆pyrF∆rpoZ+pSR01 growth results:
- When no IPTG was present, the bacteria grew in LB+amp, NM+his, and NM, showing that the plasmid rescues the selection stain's growth phenotype! This implies that our strain had the right genotype all along. However, when IPTG was added to the media, the strain only grew well in LB. This is very odd, since IPTG was supposed to induce zif268 and thus His3 expression. We will try a couple things to make sure nothing went wrong:
- redo cultures in the same pattern as yesterday--perhaps IPTG+/- were mislabeled
- inoculate cultures of ∆HisB∆pyrF∆rpoZ +/- pSR01 so that Vatsan can use a plate reader to compare their growths
- Joung wrote back and said that the HisB deletion is in fact 6 bases in frame. He also mentioned something about a lac deletion which we are looking into.
MAGE3 sequencing results:
- none of the colonies had the stop codons. This may be because mutS is not knocked out, and it can correct strands that have up to 5 bases mismatched.
Kan-ZFB for lambda red:
- Noah did lambda red in EcNR2 and the plate was covered in crystalline patterns that may be cells. We will test this by:
- Restreaking some of these "colonies" onto a fresh kan plate to get the bacteria more spread out
- Streaking a kan plate with pKD46 (from glycerol stock) to see if the kan is effective
- Taken a streak off the plate, diluted it in 200µL H20, and used 1µL as template for PCR
- KAPA, 1529620-flanking primers, 90 sec elongation and 56 anneal, 25 cycles
- Results: 300bp band--the streaks on the plate are bacteria, but they do not have the insert.
- We also retried the thio kan-ZFB PCR using some new primer aliquots. Once again it failed.
- Because this PCR keeps failing, we're going to try a different way and do two separate and shorter PCRs to be joined later in an overlap PCR:
- PCR of kan cassette: kan cassette as template (diluted 6/16 aliquot), kan F phosphothio and kan R primers, KAPA, 56˚C anneal and 90 sec elongation
- PCR of ZFB-wp-hisura: ZFB-wp-hisura as template (diluted 6/16 aliquot), ZFB-wp-f and URA3 R phosphothio primers, KAPA, 60˚C anneal and 90 sec elongation
Team TolC
- PCR ECNR2
- Ran PCR on four colonies from lambda red recombination results, ECNR2 liquid culture from Church lab, and colony from original ECNR2 plate
- Grew each up in LB Amp liquid culture
- Tomorrow the gel will tell us whether the insertion was successful and also if the ECNR2 cultures are pure
- Ran PCR on four colonies from lambda red recombination results, ECNR2 liquid culture from Church lab, and colony from original ECNR2 plate
- PCR Noah's MAGE on rpoZ knockout
- Picked 15 colonies from each plate with ECNR2 without TolC and ECNR2 with TolC
- Created liquid cultures of all 30 colonies picked and growing up over night
- PCR will be ready to send out tomorrow for sequencing
- Picked 15 colonies from each plate with ECNR2 without TolC and ECNR2 with TolC
- More kan-ZFB-wp insertion construct needed
- Ran PCR in order to obtain more kan-ZFB-wp insertion construct from the kan-ZFB-wp-His3-PyrF construct
- Placed on gel and used gel extraction in order to purify
- 10.7 ng/
Team Web Design
- Safety, safety, safety!
- Received safety confirmation letter
- Finalized project description
- Updated our iGEM wiki page for the upcoming deadlines (Project description and safety, due July 15th)
July 14th
Team ZF
Results of the expression plasmid transformation
Only samples 78 to 82 had colonies after overnight incubation--these were the same colonies that were electroporated, instead of chemically transformed. Because we only added 1 ul of our cells to the plates, it is possible that this was not enough sample for us to see colonies, especially since chemical transformation is not as efficient as electroporation. The samples on the LB+spec plates had what appeared to be unspecific growth around the edges. This may be because the spec did not diffuse evenly. We will be re-plating all of our samples later today, on new LB+spec plates.
PCR of product from isothermal assembly of expression plasmid
We also ran a PCR on the product of the three-part isothermal assembly (omega+linker, ultramers, spec backbone). We used the cross-junction primers and the same recipe as our PCR on Tuesday. Once again we expected a product of 1.4 kb.
There were fairly sharp bands underneath the band we expected. Additionally, the expected 1.4 kb band was not visible for samples 80 and 81, although they were successfully grown on plates. Sample 85 had no bands, not even primers.
Since 80 and 81 grew on our plates yesterday, we continued with our plan of re-plating our original transformations, especially since the chemically transformed cells do have the appropriate isothermal assembly product to insert. We re-plated all the transformations on the LB+spec plates made today, and will leave them overnight in the 37* incubator. We also made cultures from the colonies on plates 78-82 to mini-prep tomorrow.
Rerunning of PCR products (ultramers and isothermal assembly)
In addition, we decided to see if the streaking was possibly caused by the gel instead of a PCR problem, and so we ran three of our products in a 1% agarose gel instead of an E-gel, with the results below.
Team Web Design
- Intro video: brief (~1:30) Flash video explaining our project (Our project in 30 sec? 1 min?)
- Interview Joung/Persikov-- videotape (documentary style) for Human Practices page
- Human Practices
- Broader societal impacts
- Gene therapy, personalized medicine
Team Wolfe
thio kan and thio ZFB-wp-hisura PCR:
- Ran overnight PCR of kan cassette and ZFB-wp-hisura on E gel: looks great!
- PCR purification with Qiagen kit:
- added an additional 300µL buffer PB to adjust the pH
- thio kan cassette: 59.9ng/µL 260/280=1.90
- thio ZFB-wp-hisura: 100.9ng/µL 260/280=1.89
- Overlap PCR:
- 4 rxns: two used undiluted template (60ng thio kan cassette and 101ng thio ZFB-wp-hisura), two used template diluted 1:10
- KAPA, 56˚C anneal, 90 sec elongation, 25 cycles
- primers (kan f phosphothio and ura3 r phosphothio) added after 10 cycles
- Results: PCR worked but with tons of side products so we will try to optimize the procedure. Expected product = 3Kb, notable side products = 2Kb, 4Kb. N.B. The samples were accidentally loaded while the gel was running so they are staggered on the gel.
- Gel stab PCR
- DNA from the 3Kb e-gel band were removed using the stabbing technique and placed in 4 respective reaction tubes (diluted in 20µL water)
- The 4 samples were separated into template dilutions of 20X, 100X, 200X, and 400X and phosphothio primers for PCR to amplify the desired 3Kb kan-ZFB-wp-hisura sequence. PCR results will be reported tomorrow.
- Template PCR followed by PCR with primers:
- PCR just the template (1µL diluted thio kan cassette and 1µL diluted thio ZFB-wp-hisura) with no primers, KAPA, 12 cycles, 60˚C anneal, 90 sec elongation
- take PCR product and use it as template for new PCR either undiluted, diluted 1:4, 1:16, 1:64, 1:256; kan f phosphothio and ura3 r phosphothio primers, 25 cycles, 59˚C anneal, 90 sec elongation
pSR01 phenotype:
- Just to be sure, we will assemble cultures in a flat-bottomed 96 well plate and use a plate reader to track the growth curves.
- 2 colonies ∆HisB∆pyrF∆rpoZ, 10 colonies transformed with pSR01
- media: LB+amp, NM+his, NM, NM+3-AT (1mM), LB+amp+IPTG(100µM), NM+his+IPTG, NM+IPTG, NM+3-AT+IPTG
- plate reader: absorbency of 600nm, measurements every 10 min, shaking 560 sec before each measurement (settings saved in iGEM folder on computer in tissue culture room)
Team TolC
- PCR done of 4 colonies picked from KAN plate (with ECNR2 after recombineering with the KAN-ZFB-wp construct. Running an E-gel showed that the insert was not present. Since the KAN plates were old, it might be speculated that the plates were old, and so, the colonies were not selected for KAN resistance.
- Primers were TolC_seq_F and TolC_seq_R.
- Without insertion these two primers will create 449 bp sequence, and with insertion length will be 1650 bp
- Primers were TolC_seq_F and TolC_seq_R.
- Some of the original culture after recombineering, 1ml solution spun down, and resuspended in 100μl of LB, and plated on new KAN plates. To check, one of the old KAN plates was plated the same way and put in at 2:30pm. (These plates will be checked tomorrow morning.)
- PCR done of a few of the rpoZ MAGE with stop codon insertions. Oligo name: rpoZ_MAGE_KO (Noah's oligo) (Noah's plates, after two cycles of MAGE, on both ECNR2 and ECNR2ΔTolC). The side product in the gel could be due to a low annealing temperature of 53°C, so we should do a gradient PCR (e.g.55-66°C) to get the most product the next time.
July 15th
Team Wolfe
Thio kan-ZFB-wp-hisura optimizing PCR results:
- The gel stab PCR produced a product of the right size, but the 2kb side product was still brighter. The 2-PCR protocol also still had substantial side products but at least they did not completely overshadow the real product. We will accordingly go ahead and do a gel extraction on the 2-PCR samples so that we can try lambda red with them next week.
pSR01 growth phenotype results:
- Unfortunately the computer hooked up to the plate reader spontaneously restarted during the night to download updates, so we lost all the data. Just by looking at the plate, it still seems like IPTG is inhibiting growth. The ∆HisB∆pyrF∆rpoZ controls also did not grow, probably because the plate the colonies were taken from was too old, so we will restreak from the glycerol stock and set up the plate reader again this weekend.
Team ZF
PCR of electro comp expression plasmids
We performed a mini-prep of the cultures that we grew overnight of the electrocompetent cells. We then performed a PCR on the product of the mini-prep, checking for the 1.4 kb cross junction to confirm the presence of our omega+ultramers+spec plasmid. We used the same recipe as the PCR we performed on Tuesday.
No bands appeared for PCR peformed on a 1 to 10 dilution of the cultures. Miniprep product from colonies 78, 79, and 82 had the expected band at 1.4 kb, while 80 and 81 had a band that was slightly smaller than the the expected product size. We saw a similar band in our PCR of the isothermal assembly reaction product. We are unsure exactly what is causing this band. If either the omega subunit or ultramers were missing from the plasmid, a similar sized product would result since both are ~350 bp. We are curious if a plasmid can still be formed if one of these components were missing.
We checked the concentrations of our miniprep product to see if that would explain our problem.
ID Conc. (ng/ul) 260/280 1.4 kb band? 78 16.12 2.10 Y 79 41.64 1.64 Y 80 5.84 2.01 N 81 61.24 1.62 N 82 20.25 1.72 Y
We can't tell whether the concentration of the miniprep product contributed to the strange band we saw. 81 had the highest concentration but still didn't work. We decided to try plating 4 more colonies from 80 and 81, to see if any of them have the appropriate band.
We also decided to make final plates of cultures 78, 79, and 82, using 2 ul of culture and 50 ul of LB+spec. They were plated on new spec plates. We will incubate them at 37* overnight.
Checking the expression plasmid plates
All the plates had colonies on them, although there were still areas of unspecific growth, presumably because of uneven dilution of the spec into the LB plates.
We made cultures of all the chem comp cells (77, 83, 84, 85) and the electro comp cell that did not have the correct insertion (80, 81). We picked 4 colonies from each plate, resulting in a total of 24 cultures. We have left them in the 37* shaker overnight, and tomorrow plan on performing a miniprep and PCR to check for the cross-junction.
PCR to check for omega+ultramers
We performed a PCR on the five electro comp miniprep products, to find out what may be missing. We used the omega_F+pLacO (index: 73) as our forward primer, and each of the respective reverse ultramers as our reverse primers (index: 78-82). We had an annealing temperature of 58* and an extension time of 30 seconds, and the expected product size is 646 bp.
July 16th
Team ZF
Colony minipreps
Minipreps were performed on all of the samples except for a few where nothing grew in the LB/spec media. These exceptions included 85.3, 85.4, 84.2, 81.2, and 81.3. Possible reasons for no growth for these samples may include: (1) The colony was somehow not a transformed colony and was a cheater (possibly picked from a spot where there was a smaller amount of spec in the unevenly distributed dish), and (2) the colony did not get picked up properly with the pipet tip.
The concentrations of the DNA after miniprep were somewhat low, and a full table of these concentrations and DNA purities can be found below:
PCR on minipreps to check cross junction
PCR was done on the miniprep products to check for the cross junction length, expected band size of ~1.4 kb. The protocol used was the same as the previous time. In addition to PCR-ing the miniprep products, a positive control was also included, which was the finished Zif268 + omega + spec backbone plasmid (this was the first successful junction PCR we obtained, and so it should work again).
Replating final samples for 78, 79, 82
Strangely, nothing grew on the plates that we seeded yesterday with 2ul of bacteria. We seeded plates again for samples 78, 79, and 82 with 15ul of transformed bacteria instead, and hopefully these should grow by tomorrow.July 17th
Team ZF
Replating final samples, take 2
We realized that the plates we seeded yesterday (July 16) were seeded from the transformation bacteria tubes, which has a mix of bacteria that may/may not have taken up the plasmid. These plates therefore cannot serve as the final stock plates. We took the E. coli from our initial colony picking and put them in LB + spec to grow to saturation tomorrow, where we will replate again with those bacteria (for which the good PCR results should correlate).
Gel analysis of miniprep junction PCR
The miniprep junction PCR products from yesterday were ran on an agarose gel, with the results below:
July 18th
Team ZF
Sending expression plasmids out for sequencing
We decided to send out the expression plasmids that had bands at approximately the correct size out for sequencing. We originally wanted to sequence the plasmids, but our miniprep concentrations were not high enough. Apparently, we should be getting high yields from minipreps, but thus far our yields have been around 50 ng/ul at the highest.
We decided to sequence the unpurified PCR products instead. In order to do this we had to re-do the PCRs for Zif268, FH bottom (78), CB top (79), Myc 981 (82). Below are our 1% e-gel results, verifying that we had appropriately sized bands:
Our final samples sent out for sequencing are:
Index Sample ID Index Sample ID 1 77_1 11 80_3 2 77_2 12 80_4 3 77_3 13 - 4 77_4 14 83_2 5 84_1 15 83_3 6 84_4 16 83_4 7 81_1 17 Zif268 8 81_4 18 FH_bottom 9 80_1 19 CB_top 10 80_2 20 Myc_981
Making more cultures and plates of our expression plasmids
We picked two more colonies from plate 85 today and made 5 ml cultures from them. We also made 2 fresh cultures from the old 85 cultures, by taking 50 ul of old culture and 5 ml LB+spec. We will leave them overnight in the 37* shaker and perform minipreps and PCRs on them tomorrow.
We made positive control cultures from 81.1, 81.4, 80.1, and 80.2, in order to test our minipreps tomorrow. They reactions have a wide range of concentrations post-miniprep, and we are curious to see if the same will happen after performing a miniprep a second time.
Another way to figure out if our expression plasmids contain the appropriate insert is by using a restriction enzyme to cut it and then performing a PCR on the linear DNA. We had previously inserted 2 cutsites on our plasmids, Bsa1 (for insertion of finger 1) and Xba1. The lab already has Xba1, and we ordered Bsa1. Xba1 can be affected by methylation if it is preceded by 'GA' or followed by 'TC' [http://www.neb.com/nebecomm/products/faqproductR0145.asp#874]; we checked and our sequences should not be affected.
We also re-plated 78, 79, and 82 today, one set of plates with 1 ul of culture, and the other set with 10 ul.
Creating the 96 well test plate
Team Wolfe
SNOWMAN!
EcNR2 lambda red (Noah's prep):
- Noah remade the kan-ZFB-wp-hisura insert for us with the kan cassette in the opposite orientation from the his3 and ura3 genes. (The kan cassette is under a strong promoter and does not have a transcriptional terminator, so it's possible that RNA polymerase might keep transcribing through the end of kan and into his3.) He also used it for lambda red in EcNR2, and a couple of plates look like they may have a couple colonies.
- picked 4 colonies, diluted in 20µL H20 and used 1µL as template for 1529620 locus PCR. The rest was grown in LB+kan.
Lambda red with Noah's kanZFBhisura:
- We recently discovered that the kan plates we had been using were very old, and that might be why we got that weird crystalline lawn of colony-like things. We will retry lambda red in the ∆hisB∆pyrF∆rpoZ +pKD46 strain and see if this solves the problem
- 1.5mL cells divided in two: half for kan-his-ura (300ng) and half as a negative control (water)
Thio insert:
- In case lambda red also does not work, we will try to add phosphothioate bases to the insert, but only to the lagging strand 5' end (so only the Ura3 side). Since our primers will have very different melting temps, we will try 2 different reactions:
- Touchdown PCR:
- template: Noah's kan-ZFB-hisura (163ng/µL) diluted to ~10ng per reaction (4 reactions)
- primers: kan_f+homology, Ura3 R phosphothioate
- 2 cycles at each annealing temperature (67, 66...57) for 11 cycles followed by 15 more cycles at 57; 3 min elongation
- Gradient PCR:
- same as above but with 12 reactions, annealing temperatures from 57-67˚C
- Results: most of the gradient reactions produced a faint but clean band at 3kb. The touch down PCR had a stronger 3kb band but also lots of side products.
Team TolC
Primers
- Created and ordered primers in order to test the presence of MAGE rpoZ knockout oligo
- Primers made are named
- rpoZ_MAGE_KO plus 4, rpoZ_MAGE_KO minus 4, which have all 4 of the stop codons
- rpoZ_MAGE_KO plus 2, and rpoZ_MAGE_KO minus 2, which have only the first two stop codons.
- Primers made are named
Kan Plates
- Performed control experiment on Kan plates by growing up liquid culture from glycerol stock and then spreading on Kan plate in order to determine the speed of growth and intensity of the presence of Kan on the plates
- Will check on these tomorrow.
Kan-ZFB-wp insertion
- Picked 5 colonies from ZF090 and ZF091 plates for a total of 10, ZF092 had too few colonies to pick from
- Placed colonies in 20 μL of water and used 1 μl for a 20 μL PCR reaction
- Then took 10 μL from 20 μL dilution and added to 100 μL of LB in 96 well plate and put in 30°C fridge, at 3:19 pm
- Same protocol as KAPA PCR with the following changes made
- Step 3, gradient for annealing, 55°C - 77°C, for a range of 55,57,59,61 and 63°C for the two rows of 5 PCR tubes each (placed leftmost)
- Step 4, Extension, 1 min
- Step 5, 25 cycles total
- Name: TOLCGRAD, PCR5 machine.
July 19th
Team ZF
Checking on plates 78, 79, 82
Both the 10 ul and 1 ul essentially had a lawn of bacteria growing on them. We re-plated these samples, using 1/100 ul and 1/10000 ul of bacteria in 50 ul of LB. We will leave these plates to incubate overnight at 37*.
We also made a culture from the old 85 culture, which we will plate tomorrow.
Minipreps of colonies and controls
Our minipreps from Saturday had low yields, especially for culture 85. We suspect this is why the PCR [[#Gel analysis of miniprep junction PCR on Sunday did not work for this sample. Unfortunately, 85.3 and 85.4 did not grow even after being left in the shaker overnight. These were two cultures we made by picking new colonies from plate 85. This may have occurred because we did not actually pick the colony, or the colony was a cheater. Thus, we performed minipreps on 80.1, 80.2, 81.1, 81.4 (the controls, chosen for their wide range of concentrations after our first miniprep), and 85.1 and 85.2.
In order to figure our why our minipreps were resulting in unexpectedly low yields and increase our yields:
- We tested the pH of our water and Buffer EB.
- The pH of our aliquot of water was ~4-5 (too acidic...we are unsure why)
- The pH of our Buffer EB was ~7.
- Ideally, the DNA should be eluted by a solution of pH 7, so we decided to use Buffer EB during the elution step.
- We also checked the copy number of the origin of replication used in our plasmids. ColE1 apparently has a low copy number, which might also explain our low yields.
- We had made 5 ml of culture in an attempt to increase our yields.
Our yields were larger than our first round of minipreps, although the purities were rather low. We decided to proceed anyway.
PCR of miniprep products
We then ran a PCR for the cross-junction on all 6 (80.1, 80.2, 81.1, 81.4, 85.1, 85.2) samples, using our usual cross junction protocol.
We saw bands for samples 80.1, 81.1, 81.4, 85.1, and a very faint band for 85.2. The strange thing about these bands is that they appeared to be right around 1650 bp, larger than our 1.4 kb target size. We are unsure of the identity of these bands and will send them out for sequencing.
Starting a new plate for sample 85
Since we have been having such bad luck with sample 85, we seeded 2 ml of LB+spec with 100 ul of the transformed bacteria for sample 85. This is in preparation for making a new plate of sample 85 (the old plate was not a good spec plate and had lots of unspecific growth as well as very few colonies, so if we used this plate for more colony picking our choices would be very limited). This new plate should also give us more luck with picking colonies and having them actually grow in the LB+spec.
Designing ultramers for Zif268, OZ052, and OZ123
In preparation for designing our 96-well practice plate, we designed ultramers for our three positive controls Zif268, OZ052, and OZ123. These ultramers contain the Type II binding sites as well as the F2/F3 fingers. Since the practice plate will contain oligos that encode for F1, we can use these three positive controls to practice and ensure that we secure the technique of using the Type II binding sites to swap in F1 into our expression plasmids. The ultramers have not yet been ordered.
Team TolC
PCR to test insertion of KAN-ZFB-wp into ECNR2
- Made 26 dilutions of culture from three different recombineering plates
- 10 cultures from Kan_tolC_locus_F1 (ZF090 plate) primer recombination
- 10 cultures from Kan_tolC_locus_F2 (ZF091 plate) primer recombination
- 5 cultures from Kan_tolC_locus_F3 (ZF092 plate) primer reecombination
- 1 culture from an old Kan plate with recombination from last week
- 5 of ZF090, 6 of ZF091, and 5 ZF092 were placed in a PCR with 58˚C annealing temperature
- 5 of ZF090, 4 of ZF091, and 1 from old plate were run in a PCR with 62˚C annealing temperature
- primers used were TolC_seq_F and TolC_seq_R
- Chose colonies based on the intensity of the bands and grew up cultures of ZF091-2d, ZF091-4d, ZF090-1d, ZF091-5d, ZF090-1s, ZF090-2s, ZF090-3s, and ZF091-3s
Sequencing of Kan-ZFB-wp insertion
- Prepared 16 PCR products of the 8 cultures we chose to grow up in order to send out for sequencing
- Used TolC_Seq_F and ZFB-wp-F primers
- Bands looked a little small when ran out gel for longer
New MAGE oligo
- Constructed a new oligo for MAGE knockout of rpoZ, with oligos more than 4 mutations and substitutions the efficiency drops from 15% down to below 5%, so we created a rpoZ knockout oligo with only 2 substitutions
- The new MAGE rpoZ knockout is rpoZ_MAGE_Subs
- Also ordered primers to test the presence of the knockout rpoZ_MAGE_Subs1+, rpoZ_MAGE_Subs1-, rpoZ_MAGE_Subs2+, rpoZ_MAGE_Subs2-
Team Wolfe
EcNR2 lambda red colony PCR results:
- PCR showed that colonies 1-3 had the insert while 4 did not. Yay! (Note that in the gel image, we accidentally cut off the 300bp band in lane 4)
- We will grow up a reinoculated culture of colony 1 for a glycerol stock. If the Wolfe strain keeps not working, we at least have the insert in an EcNR2 line and can hopefully MAGE out the other genes without too much difficulty.
pSR01 growth phenotype:
- We will try to used the plate reader again to compare the growth phenotypes of ∆HisB∆pyrF∆rpoZ and ∆HisB∆pyrF∆rpoZ+pSR01. Used the same media as last time but since we're using a 48 well plate, each well has 250µL media. We used 5 pSR01 colonies and one selection strain colony as a control.
- N.B.: this plate we are using is not necessarily meant for OD tests like this and may give some variable readings.
Lambda red results:
- Yesterday's lambda red, which used Noah's kan-ZFB-wp-hisura, has yet to produce colonies. We will leave the plates in longer in case any come up later.
Thio kanZFB:
- Since yesterday's lambda red may not have worked, we need to get the thio kan up and running. We ran out the touchdown PCR samples (since they have more DNA) and will gel extract.
- 57.7ng/µL 260/280=1.98
- 69.3ng/µL 260/280=1.98
July 20th
Team ZF
Checking on 1/100 ul and 1/10000 ul 78, 79, and 82 plates
We got colonies on all the plates, but the 1/100 ul plates look a bit crowded. On the other hand, the 1/10000 plates only have a few colonies. We decided to save both plates, and decide which to use later.
- Based on these results, a 1/1000 ul dilution might be best.
Check on 85 culture
There was no growth in our culture.
Later: It ends up that our ultramer from 85 (Pos Con 77) had an error in it. Instead of coding for fingers 2 and 3, it codes for fingers 1 and 2. We ordered a new ultramer, and we no longer need to work on 85.
Finalizing the practice plate
We decided on using a practice plate with 24 oligos, consisting of 18 target sequences, and 6 controls. Our 6 controls are Zif268, OZ052, OZ123, and 3 sequences from the CODA table. We will need to construct the expression plasmid for Zif268, OZ052, and OZ123. Their ultramers were ordered today.
Our 18 targets were GNN/TNN sequences, which are more likely to have a binder, and were distributed among the backbones. The target and control groups each have their own set of primers, to facilitate picking them from the pool.
Expression Plasmid Sequencing Results
We got back our sequencing results:
| Name | ID | Final? | Notes |
| FH Top (77) | 1 | N | 1 snp |
| 2 | N | 5 snps | |
| 3 | Y | G -> C. Still Ala | |
| 4 | N | 1 snp | |
| FH Bottom (78) | 1 | N | 1 Deletion at 1746, last nucleotide of F of FQRICMRN |
| CB Top (79) | 1 | N | A lot of deletions/mistakes in f2/f3 region |
| CB Bottom (80) | 1 | Y | |
| 2 | Y | G->C. Stiil Ala | |
| 3 | N | deletion 1613-1631 | |
| 4 | N | many mistakes at end, may be ok | |
| Myc 198 (81) | 1 | Y | |
| 4 | Y | G->C. Still Ala. | |
| Myc 981 (82) | 1 | Y | |
| Pos Con 16 (83) | 4 | Y | G->C. Still Ala. |
| 2 | N | A lot of deletions/mistakes in f2/f3 region | |
| 3 | N | A lot of deletions/mistakes in f2/f3 region | |
| Pos Con 55 (84) | 1 | Y (?) | Right sequence, but messy trace |
| 4 | N | at 547: deletion at type II binding site
at 665: deletion at beginning of F3 | |
| Zif 268 | 1 | Y |
| Summary: |
| Good: 77.3, 80.1, 80.2, 81.1, 81.4, 82, 83, 84.1 (?), Zif268 |
| Bad (Redo): 78, 79 |
We picked 4 new colonies for 78, and 79 since their sequences had many errors. We left these cultures overnight at 37* in the shaker.
Construction of OZ052 and OZ123: Joining the Finger with Homology regions
Previously, we did a PCR to reassemble the ultramers for OZ052 and OZ123, but they only coded strictly for F1/F2/F3 and did not have any overhang homology with any surrounding code. Thus, we performed PCR to add these homology overhangs so that we can integrate them with the omega subunit as well as the spec backbone to create a final expression plasmid.
The program we used was 55* annealing temperature, and a 15 second extension time. Gel results pictured below.
We had an expected product size of 400 bp., which we can see in the gel. Bands are a little weird since there are bands heavier than the heaviest theoretical band (which is the strong product). We will do a gel extraction tomorrow to get rid of these bands to obtain pure product for use in the isothermal assembly.
Team TolC
rpoZ MAGE knockout
- Ran two rounds of MAGE on 4 ECNR2 cultures with the kan-ZFB-wp insertion using Noah's rpoZ knockout oligo
- Grew cultures up to mid-log in kan+amp and LB
- Four cultures are ZF091-2d, ZF091-4d, ZF090-1d, and ZF090-5d
- Electroporator read 5.4 ms for all four cultures in first round
- Could not do a third round of MAGE since the E.Coli do not appear to have recovered, and are nowhere near midlog (MAGE round 2 was done by 5:15pm and the cultures were checked at 8:30pm). Will let the culture go on overnight.
- Put 50ul of a 1:100 dilution of each culture on Amp plates. Used a spreader for plates 1&2 and glass beads for plates 3 & 4.
Glycerol Stocks
- Made 4 glycerol stocks of the cultures sent to be sequenced
- ZF090-1s, ZF090-2s, ZF090-3s, ZF091-3s
convinced that the gradient PCR machine is out of order, ought to inform Andrew
Team Wolfe
IPTG Plate and the Plate Reader
- Technical delays have allowed us only a few hours worth of plate reading for a 46-well plate with selection strain + pSR01 in LB+Amp +/- IPTG, NM+His +/- IPTG, NM +/- IPTG, and NM+3-AT +/- IPTG. Strains used: ∆HisB∆pyrF∆rpoz+pSR01 (5 colonies) and ∆HisB∆pyrF∆rpoz (1 colony).
- The plate had a noticeable white residue under the lid around around the edges. We believe this was due to flaking of the plastic cover from vibration in the plate reader.
- Cell growth in the plate suggested that IPTG is killing our cells since few cultures survived with IPTG and those that did (LB+Amp) grew less than the equivalent cultures without IPTG.
New Plate Without IPTG
- On plate reader at 11:30AM
- Strains used: Water as control, Selection (Wolfe) Strain, Selection Strain + pSR01
- Media Used: LB+Tet (LB+Amp for SS+pSR01),NM+His, NM, NM+3-AT (1µM), NM+3-AT (2.5µM), NM+3-AT (5µM).
Lambda Red Same procedure as usual
- Added ~350ng of Thio-Kan-ZFB-wp-His3-Ura3
- Used "new cuvettes"
- Negative water control
Plate Reader Update TEMPORARILY OUT OF SERVICE
Your worst nightmare
http://www.youtube.com/watch?v=v4o8MDjuu-AJuly 21st
Team ZF
Checking colonies 78 and 79
All of the cultures grew overnight, so we performed a miniprep on them to obtain the plasmids.
Our concentrations were better than they have been in the past; notably, after the addition of buffer P2, our solution actually turned a bright blue (indicating lysis of the cells), which we had not seen before.
We then ran a PCR on our miniprep product, to check for our desired insert. We used our usual protocol for a PCR of the expression plasmid cross-junction.
Some of the products did not show up very strongly, but we sent them out for sequencing in case it was a PCR/gel error.
Sending out our expression colonies for sequencing
We sent out the samples from yesterday that had no more than 1 SNP for reverse sequencing. In addition, we also sent out 78 and 79 for forward and reverse sequencing.
- Reverse sequencing: 77.1, 77.3, 77.4, 78.1, 80.1, 80.2, 80.4, 81.1, 81.4, 82.1, 83.4, 84.1, 84.4, Zif268
- Forward and reverse sequencing: 781, 78.2, 78.3, 78.4, 79.1, 79.2, 79.3, 79.4
For sequencing, we used the forward primer pZE23G_3581_F CCAACCTTACCAGAGGGCGC (Junction forward) and the reverse primer pZE23G_2123_R CGAACGACCTACACCGAACT (Junction reverse)
We did a PCR of the sequences from yesterday that we want sequenced again.
The bands were clear, and we will send these products to be sequenced.
Gel Extraction and Isothermal Assembly of OZ fingers+homology
We ran a gel extraction of our PCR product from yesterday, so that we could use it for isothermal assembly later. Strangely, we got bands that were larger than our largest theoretical product.
We had the following yields from our gel extraction:
ID Conc. (ng/ul) 260/280 OZ052 96.3 1.93 OZ123 125.5 1.91
Based on these concentrations, we used the following recipes for our isothermal reaction:
- OZ052:
- Spec: 0.63 ul
- OZ052: 1.56 ul
- Omega: 0.13 ul
- Water: 1.56 ul
- Isothermal assembly solution: 7.5 ul
- OZ123:
- Spec: 0.63 ul
- OZ123: 0.14 ul
- Omega: 0.13 ul
- Water: 1.60 ul
- Isothermal assembly solution: 7.5 ul
We left the isothermal assembly product in the -20* freezer, and will perform the transformation tomorrow.
Team Tolc
Team Wolfe
- Lambda red culture did not grow colonies overnight on a kanamycin plate. This may be due to a low time (4.8ms) during the electroporation.
- Repeated lambda red using the same procedure. Got a similar time constant and noticed that the water negative control had more cells than the culture containing the insert, but we went ahead and plated 1mL of each on kan plates.
- Also replated the 7/20 lambda red culture to see if we can get any good cells (1.5mL).
July 22nd
Team ZF
Sequencing Results of Expression Plasmids
The good news is that we managed to get a perfect sequence for each of our plasmids. The bad news is that we do not know which sequences are the matches, since the names are all scrambled for some reason (i.e. sample 78.x does not align with FH bottom like it should, but instead aligns perfectly with another plasmid sequence). We will thus need to resend our samples for sequencing to be able to choose the correct plasmids that match. Below is a summary of the scrambled data from the sequencing we sent in yesterday:
| ID | Is a perfect match with | Notes |
| Myc 198 (81) | 78.2, 78.3 | |
| FH Top (77) | 81.4, Myc 981 | 83.4 has 1 deletion |
| CB Bottom (80) | 84.4 | 78.1 has 5 deletions; Zif has 1 deletion |
| FH bottom (78) | n/a | 84.1 deletion at beginning of F3 |
| CB Top (79) | 79.3 | |
| Myc 981 (82) | 78.4 | |
| Zif 268 | 79.4 | |
| Pos Con 55 (83) | 79.2 | 79.3 has deletion at beginning of F3 |
| Pos Con 16 | 79.1 |
To this end, we redid the PCRs for everything in preparation for the resequencing, using the junction primers and the previous protocol. The resulting gels can be observed below:
From these PCRs, all the reactions look like they produced bands of the right size, a good indicator that the isothermal assemblies worked. Only sequencing will tell whether they are truly the products that we are looking for.
We sent these samples out for sequencing, making sure that all the samples were in the appropriate tubes.
Transformation for OZ plasmids
Today, we took the results of the isothermal assembly from yesterday and did chemical transformation to introduce the plasmids into the Top 10 ChemComp bacteria, using our protocol for chemical transformations. They were left to recover for approximately 1.5 hours. We plated 2 dilutions, 10 ul and 50 ul, and left them overnight at 37* in the incubator.
Team Wolfe
- Yesterday's lambda red plate had several large colonies--but so did the negative control plate. We will go ahead and do a PCR of the 1529620 locus (as previously described) on 12 of the colonies in case one of them happens to have the insert. The colonies were also grown up simultaneously in LB+kan.
- Results: all of the colonies were cheaters
- In case these colonies do not have the insert, as we suspect, we will repeat lambda red with the thio kan-ZFB-wp-his3-ura3 insert
- 1mL culture for each condition, 210 ng DNA
- time constant for thio insert sample was 5.0
Team TolC
Gel for Jun's MAGE rpoZ, rowD
- The gel appeared to have bands, so we are running a second PCR to confirm it
July 23rd
Team Wolfe
- Lambda red results: no real colonies, only cheaters.
July 24th
Team ZF
Sequencing Results
We got back our sequencing results from the samples we sent out on Friday, and the results are summarized in the table below. The checkmarks indicate whether the sample is adequate for usage or not.
| FH top (77) | Notes | |
| ✓ | 77.1 | Only G -> C in ala |
| ✓ | 77.3 | Only G -> C in ala |
| 77.4 | Single deletion in F3 | |
| FH bottom (78) | ||
| 78.1 | Many, many SNPs throughout, oddly F and R differ | |
| 78.2 | Many, many SNPSs, R starts in a strange place | |
| 78.3 | Four deletions throughout on both F and R | |
| 78.4 | Many, many SNPs throughout | |
| Original | Only one SNP in F3, but unacceptable | |
| CB top (79) | ||
| 79.1 | Many, many SNPs and deletions throughout, F and R the same | |
| ✓ | 79.2 | No SNPs |
| ✓ | 79.3 | No SNPs |
| 79.4 | G -> C in ala, deletion in the Q of the F3 \"FSQ\" | |
| CB bottom (80) | ||
| ✓ | 80.1 | No SNPs |
| 80.2 | G -> C in ala, deletion in the homology region shortly after F3 ends | |
| 80.4 | G -> C in ala, five deletions in F3/homology region | |
| Myc 198 (81) | ||
| ✓ | 81.1 | No SNPs |
| ✓ | 81.4 | Only G -> C in ala |
| Myc 981 (82) | ||
| ✓ | 82.1 | Only G -> C in ala |
| Pos con 16 (83) | ||
| ✓ | 83.4 | Only G -> C in ala |
| Pos con 55 (84) | ||
| ✓ | 84.1 | Only G -> C in ala |
| 84.4 | G -> C in ala, two deletions | |
| Zif268 | ||
| ✓ | Zif268 | No SNPs |
July 25th
Team TolC
Sequencing of Jun's rpoZ MAGE knockout
- Prepared six products and primers for sequencing
- Sending Jun colony 4, PCR's 1-mutant_R, 2-mutant_R, 3-mutant_R, 4-mutant_R, 2-mutant_Int_R, and 4-mutant_Int_R
- delayed until tomorrow (July 25, 2011) because pickup for the day already done.
Lambda Red Recombination of zeocine into TolC(assistance from Naomi, Cub team member)
- PCR out zeocin cassette (replacing the rpoZ, for selection) from Wolfe Strain
- Used primers rpoZ_F and rpoz_R, expecting a band about 1500bp? Total length about 939bp, including the promoters, according to http://rothlab.ucdavis.edu/drugs/zeo.html Roth lab @ UC Davis, Zeocin
- Ran 8, 25 µL solutions with KAPA
- PCR protocol(PCR 7 machine, "ZEOC")
- 95˚ C, for 5 min
- 98˚ C, for 20 sec
- 62˚ C, for 15 sec
- 72˚ C, for 45 sec
- Repeat 29 times
- 72˚ C, for 5 min
- 4˚ C, forever
- Gel extracted the PCR product NOTE. Since the gel, was so clean, we may do a PCR clean up next time, instead of running the PCR product through a gel for gel extraction. Just make sure to use these exact same parameters.
- Nanodrop
- 156.2 ng/µL and 107.6 ng/µL, each in 30ul of water (distilled, deionised- molecular grade)
- Nanodrop
- Performed lambda red recombination
- Time constant of 5.6 ms
rpoZ MAGE knockout attempt...yet again
- Used new oligo to perform a MAGE knockout of rpoZ
- Oligo named rpoZ_MAGE_Subs
- Electroporator read 5.2 ms
- Allowed for 3 hours of growth and spread on zeocine plate
Team Wolfe Cub
- Plate reader is still being repaired--we will have to wait to check the growth curves
- Since plans A and B are not working well at the moment, we will go ahead and start preparing for plan C (using a plasmid). Ordered primers that will clone out the pSB4K5 (a biobrick vector with a kan cassette, http://partsregistry.org/Part:pSB4K5) backbone with homology to the ZFB-wp-his3-ura3 construct. We will then connect the two with Gibson assembly.
- To this end, we need to use the biobrick DNA sent to us to transform chemically competent E coli. Unfortunately the 384-well plates that they are stored in could not be found because Andrew is out of town--hopefully they will turn up tomorrow.
- For the rest of the day, we joined team TolC for gel extraction, MAGE, and lambda red!
Team ZF
Result of OZ052 and OZ123 Transformation
Last Friday we plated out the transformed bacteria for the two OZ controls in 10 ul and 50 ul volumes. They both yielded colonies, but the 10 ul plates only had a handful while the 50 ul plates had around 15-20 colonies each. We picked four colonies from each plate and grew them in LB/spec so that tomorrow we can perform a miniprep and send them off for sequencing. We should know the final verdict by Wednesday regarding which ones are good or bad.
Glycerol stockmaking
We got the sequencing results back from last Friday, and took the following colonies to make into glycerol stock solutions: 77.1, 79.2, 80.1, 81.1 (x2 for both FH bottom and Myc 198), 82.1, 83.4, 84.1, Zif268. These colonies are a subset of the "good" colonies from our sequencing results from Sunday. We mixed 120 ul of bacteria in mid-log with 300 ul of 80% glycerol to make the final solution, which was stored at -80 degrees C.
Restriction enzyme test
We tested both XbaI and BsaI on the junction PCR product using colony 79.4 as a template (which was used because it was a "bad" sequence overall but had intact BsaI and XbaI cut sites). The junction PCR product is 1326 bp, and we expected to see bands of 593 and 739 bp with XbaI, and 717 and 578 bp with BsaI. Our restriction digest recipe consisted of the following per reaction:
- 1 ul 10X buffer (NE biolabs buffer 4)
- 7.9 ul H2O
- 1 ul DNA (~100 ng)
- 1 ul enzyme (~2 units)
- 1 ul BSA (100x)
The two gels we ran for this digest showed that XbaI did NOT work, which may be due to XbaI being too old. However, we did get double bands both times for BsaI. Our egel was strange because of uneven loading, and so we were unable to determine the true size of both bands, but the agarose gel we ran later showed the two bands in the right positions. The two gels can be seen below:
Ultramer Construction of Pos Con 77 and Other F2/F3 positive controls
We redid the ultramer overlap PCR for Pos Con 77, this time with the correct reverse sequence. We also assembled ultramers for Zif268, OZ052 and OZ123 (these are the plasmids that will have the type II cut sites in them along with F2/F3 only, as opposed to having the entire finger already assembled). For the Zif268 reaction, an annealing temperature of 65*C was used, and for the rest, the annealing temperature was 58*C. For all reactions, the extension time was 15 seconds.
The following primers were used for each reaction (index taken from the Dropbox primer index):
- Pos con 77 - 76, 122
- Zif268 - 123, 124
- OZ052 - 76, 125
- OZ123 - 76, 126
July 26
Team ZF
Isothermal Assembly of Pos Con 77, Zif 268, and OZs
Today we checked the PCR products of the ultramer reaction from yesterday. They did not look good. We repeated the reaction using a touchdown PCR beginning with an annealing temperature of 70 degrees and decreasing 2 degrees every other cycle until hitting a minimum of 58 degrees, and we brought the total number of cycles to 25. Extension time was 30 seconds. This looked great. The pictures for the two reactions can be seen below:
Before the isothermal assembly, we did a PCR purification and measured the concentrations of the ultramer reaction products:
We then performed an isothermal assembly, using the following mix for Pos con 77, OZ052, and OZ123:
- 0.63 ul of spec backbone at ~157 ng/ul
- 0.3 ul of ultramer (~50 ng/ul)
- 0.13 ul of omega subunit/linker
- 1.44 ul of H2O
- 7.5 ul of isothermal mix
For Zif268:
- 0.63 ul of spec backbone
- 0.19 ul of Zif (~90 ng/ul)
- 0.13 ul of omega/linker
- 1.55 of H2O
- 7.5 ul of isothermal mix
This was left at 50 degrees for an hour, then stored at 4 degrees.
Prepare control OZs for sequencing
We performed a miniprep on the eight samples for sequencing, obtaining the following concentrations:
The miniprep lysis step looked good, and the samples turned blue after lysis.
We then performed a junction PCR using the previous protocol, and checked the products on a gel:
They looked good, so we sent them out for sequencing. Hopefully we will have our final OZ control plasmids for tomorrow.
Practice Plate resuspension
We resuspended the practice plate oligos in 40 ul of water to produce a 100 uM solution, taking 10 ul of the final solutions into 90 ul of water in separate tubes to create 10 uM working concentrations. When we receive our tag primers, we should be able to test the presence of the oligos in each tube as well as amplify them in preparation for integration into the expression plasmids.
Colony finalization
For the colonies that we plated yesterday, we saw that the plates all had good growth on them and so we wrapped the plates in parafilm and put them at 4 degrees for storage. 1/1000 ul from saturated culture seems to be a good amount to plate.
Team TolC
Zeocin insertion into ECNR2
- Plated 3 mL of our recombination culture on zeocin plate, and also plated 2 mL of MAGE results on zeocin plate as a control
- The recombination plate had about 15 colonies around 9 30am, and the MAGE plate had none
- Continued by PCR of 4 colonies on recombination plate
- Each template was analysed with presence of different sized bands for rpoZ (using rpoZ_F and rpoZ_R) and presence of Zeocin cassette (using rpoZ_R and zeocin_R)
- Gel resulted with the following:
- SUCCESS!!! Yippee!!!
- NOTE. Since there was a bit of non specific annealing, we might want to use a higher annealing temperature of 64C instead of 62C
- Decided to run PCR for the Kan insertion just to make sure it was still present in the 4 colonies we chose, and it is!!!
WERE READY TO ROOOCCKKK!!!!!
MAGE rpoZ knockout
- Plated 1:10 and 1:100 dilution of MAGE results, and this morning at 10 am, they both had cells and in appropriate proportions
- If above PCR and gel of zeocin insertion does not appear to be successful, then we will run 50 PCR on 50 different colonies on the 1:100 dilution plate of MAGE rpoZ knockout
Team Wolfe
- Plate reader: trying to get in contact with Christian Daly who runs the Spectramax plate reader in the Bauer facility (Northwest basement). Hopefully we will get access soon.
- Selection system plasmid: still trying to get hold of the biobricks. Designed Quikchange primers to change the zinc finger binding sites.
- Lambda red recombination: gave lambda red one last chance, this time with a positive control
- negative control (water): plated 1mL
- positive control (kan cassette from Wyss): 50ng DNA, plated 1mL and 100uL
- thio kan-ZFB-wp-his3-ura3: 180ng DNA, plated 1mL
July 27th
- Impromptu meeting for all teams, TFs, and advisors in NW labs basement, to discuss "how we will move forward to test the selection, now that we have a TolC strain that should work for selection and a His3Ura3 plasmid-based selection that is almost complete," and to lay out and assign a lab to-do for iGEM parts, selection systems, and what to do in preparation for The Chip to arrive/backup plan in case it arrives too late (see notes in the dropbox under "Selection Meeting 7-27-11.docx")
Team TolC
Culture of TolC+Kan-ZFB-wp+Zeocin
- Did not realize that colonies needed to be grown up without antibiotic before being grown in Kanamycin, so all the three colonies from yesterday passed away and didn't make it through the night :(, so we grew up 4 more colonies and ran PCR and gel to solidify the presence of the necessary insertions
Team ZF
Checking the OZ sequencing results
| OZ052 | Notes | |
| ✓ | 1 | No snps |
| ✓ | 2 | No snps |
| ✓ | 3 | No snps |
| 4 | Deletion at F1 | |
| OZ123 | Notes | |
| ✓ | 1 | No snps |
| 2 | deletion in omega linker | |
| ✓ | 3 | No snps |
| ✓ | 4 | No snps |
We made glycerol stocks and final plates from 0Z052.1 and OZ123.1.
Transformation of Pos Con 77, OZs, Zif 268
We transformed Top 10 One Shot Electrocomp bacteria with our isothermal assembly product from yesterday. We let the cells recover for an hour, and plated 50 ul, leaving them at 37* overnight.
Team Wolfe
- Morning meeting: Since TolC has gotten their selection system onto the genome, our priorities are now going to be making the plasmid-based Wolfe selection system and getting data on the plate reader. We also will eventually try to use a genome-based selection system but using the EcNR2 strain (oligos have been ordered)
Plate Reader:
- We took a trip downstairs to B2 and were trained to use the Bioscreen plate reader in the Bauer Core facility. We now have access to a working plate reader!
- Bioscreen uses a 100-well honeycomb plate, so the organization was as follows:
- Well 1, 11, 21, etc: LB+tet
- Well 2, 12, 22, etc: NM+his
- Well 3, 13, 23, etc: NM
- Well 4, 14, 24, etc: NM+1mM 3-AT
- Well 5, 15, 25, etc: NM+2.5mM 3-AT
- Well 6, 16, 26, etc: NM+5mM 3-AT
- Well 7, 17, 27, etc: LB+tet+100µM IPTG
- Well 8, 18, 28, etc: NM+IPTG
- Well 9, 19, 29, etc: NM+1mM 3-AT+IPTG
- Well 10, 20, 30, etc: NM+5mM 3-AT+IPTG
- Column 1: no cells
- Column 2-3: ∆HisB∆pyrF∆rpoZ
- Columns 4-10: ∆HisB∆pyrF∆rpoZ+pSR01
Electrocomp Transformation
- Used Team ZF's protocol to transform electrocompetent cells with two of the pSB4K5 plasmids found in our biobricks plates (2:17H and 3:11P)
- Used 10µL ddH2O to resuspend DNA in biobrick plate wells and transferred to eppendorf.
- 2µL DNA (which, interestingly, was red...)
- plated 10µL and 100µL of each on kan plates
Quikchange primers:
- Realized that we may need to consider the bases on either side of the zinc finger binding site, so we will check this and put off ordering another day.
July 28th
Team Wolfe
Plate reader and pSR01:
- the Bauer plate reader seems to have worked very well, bu† unfortunately none of our pSR01-transformed colonies grew. This is probably because the plates were a couple weeks old. Unfortunately our glycerol stock of the strain also is missing, so we will try growing up some of the colonies in case on of them works. Otherwise, we will grow up ∆HisB∆pyrF∆rpoZ and transform again.
- Transformation: usual protocol, about 100µL DNA, recovery 2hrs, plate 1µL, 10µL, 100µL.
pSB4K5:
- The kan plates from the transformation all had colonies! We chose one from 2:17H and one from 3:11P to grow up in LB+kan for miniprep and glycerol stocks.
- Ordered quikchange primers: for OZ052 and OZ123 we added the context bases 5' and 3' of the binding sequence (found in OPEN supplementary materials). Since CODA did not include the extra bases, we left them the same as the original plasmid (i.e. Zif268's).
Team TolC
cultures of ECNR2+Kan-ZFB-wp+zeocin from F9,F11, for glycerol stocks and transformation
Sequencing
- Ran PCR and sent out sequencing to solidify the presence of the Kan+ZFB+wp and zeocin within the ECNR2 genome
- Used rpoZ_R, rpoZ_F for PCR of the zeocin cassette (and rpoZ_F for sequencing) and TolC_Seq_R, TolC_Seq_F for PCR of the Kan-ZFB-wp insert (and tolC_seq_F for sequencing)
Plasmid transformation
- Performed transformation on the Zif268 plasmid into the ECNR2 culture
- Used 3 µL in a 50 µL dilution (49+1) of 1 ul of 55 ng/µL concentration of plasmids
- 1.82 V for 5.00 ms according to the electroporator
Team ZF
Checking on Pos Con 77, OZs, and Zif 268 plates
Unfortunately, nothing grew on our plates from yesterday. Our time constants from the electroporation were rather low (~3.8), so 50 ul may not be enough to see growth. We decided to spin down the cells, resuspend them in 50 ul of LB, and re-plated this solution. We also re-did the transformation using ChemComp cells, and made 100 ul plates. Both sets of plates were left overnight at 37*.
Designing the reporter strain
Instead of placing the GFP reporter on a separate plasmid, we decided to insert it on our expression plasmid, immediately after our zinc fingers, and before the transcription terminator. We placed a ribosome binding site (Shine Dalgarno sequence: AGGAGGTT before the GFP. We will only add the GFP to Zif 268 (whole) because if we can show through GFP that the selection system works for Zif 268, we can assume it also works for the other zinc fingers. This is easier and more cost effective than ordering 12 different primers for each of our plasmids.
We will construct the reporter strain by performing a PCR to get the plasmid in linear form. We will also perform a second PCR to get out GFP and add homology regions to our Zif 268 plasmid, as well as adding the Shine-Dalgarno sequence. The primers for these PCRs were ordered today. We'll then assemble these pieces to construct a whole plasmid, and transform our bacteria.
PCR with Practice Plate
We performed a PCR to amplify our F1s from the practice plate. We used our primers for CB bottom (primer index: 66 and 67) for the 18 targets and our primers for the controls (primer index: 72 and 73) for the 6 controls. We had an annealing temperature of 52 degrees and an extension time of 15 seconds.
We got products for all of the oligos at our expected product size of 131 bp.
Glycerol stocks of successful plasmids
We happened to check our glycerol stocks, and noticed that the glycerol and bacteria were not mixed thoroughly. We remade all of our glycerol stocks and put them into the -80*.July 29th
Team Wolfe
pSR01:
- The colonies in the 96 well plate did grow overnight, but they were not saturated, implying that they were very unhealthy. We will PCR a few of them (1,2,3,6) just to see if they actually have the plasmid still
- Transformation plates look great--we will PCR 12 colonies to verify the plasmid is present
- PCR: colonies/culture diluted 1:20 as template (and colonies grown up simultaneously); M13_F and M13_R primers; 50˚C annealing and 3min elongation; 25 cycles
- Results: bands are similar to the ones seen the last time we did this PCR (and those samples had also been sequence-verified). These colonies should be good to go!
pSB4K5:
- Miniprepped both cultures. Yields weren't great (in the case of 2:17H, there actually was a spill) but they are still usable.
- 2:17H: 6.4 ng/µL 260/280=2.2
- 3:11P: 9.6ng/µL 260/280=2.4
- PCR to add ZFB-wp-his3-ura3 homology:
- 1µL of plasmid as template
- pSB4K5_F+ZFBhl and pSB4K5_R+URA3hl primers
- 58˚C anneal, 3 min elongation, 25 cycles
- Results: the product was really really really faint--primer dimers were by far the bulk of the product. We realized this is probably because one of the primers includes a Not1 restriction site, which is palindromic, 8 bases long, and all Cs and Gs.
- In order to counteract the primer dimers, we will try a gradient PCR with a range of higher annealing temperatures:
- same protocol as before except the annealing temp went from 58-65 with 8 samples (1=58, 8=65)
- Results: 3:11P did have visible product bands, but the primer dimers are still stronger
Team TolC
Sequencing of Kan+ZFB+wp and Zeocin insertions
- Sequencing results were successful and solidified presence of both insertions
Zif268 Plasmid Insertion into ECNR2+Kan-ZFB-wp+zeocin
- The plates had growth on them!!!
- So we chose 5 colonies and ran PCR on them to gel them out
- The gel shows success for colonies 2,3,4, and 5 that we chose!!!
READY FOR SELECTION!!!
Team ZF
Transformation plating results
We got no growth on any of the plated transformed bacteria for either the ElectroComp or ChemComp bacteria. To check whether the isothermal assembly itself was the problem, we ran a junction PCR using our previous protocol on the isothermal assembly mix, along with a positive control (Myc981):
As observed, there were no bands in the isothermal mix lanes while the positive control worked. Thus, we can conclude that our isothermal assembly did not work, and we will need to repeat it again (currently waiting for more isothermal mix to arrive from Wyss).
Amplification and purification of practice plate oligos
In preparation for our swap reactions (switching the oligo F1s into our expression plasmid), we mapped out the following two experiments that we will need to perform:
- Testing the selection system by preparing our expression plasmids in parallel (i.e. creating 18 plasmids in separate reactions), and then combining them all at the end and transforming them into the selection strains. This will allow us to cut and ligate the plasmids without much concern, but test whether we will be able to pick out the binder amongst the rest of the non-binders.
- Testing the cutting/ligation by combining all of the oligos first, and then carrying out the construction of the different expression plasmids all in the same tube.
Note that the results for both will ultimately yield DNA libraries if they work (except that the first experiment is much more foolproof in terms of actually yielding a library than the second experiment).
To carry out the first experiment, the following steps need to be undertaken:
- Perform PCRs on each of the practice plate oligos such that we end up with a relatively high concentration of DNA after PCR and PCR purification (over 40 ng/ul).
- Cut both plasmid backbone and oligos with BsaI type II restriction enzyme.
- PCR purify restriction products to get rid of enzyme and junk DNA.
- Run phosphatase reaction to prevent autoligation.
- Mix and ligate to get full plasmid.
- Some tips:
- Each ligation reaction should only have 10-15 ul volume.
- You should have 100 ng of backbone per reaction
- Since we are planning 18 reactions, we will need 1800 ng of backbone, which is several minipreps worth
- The backbone:oligo ratio should be appoximately 1:4
- In our case, the backbone is 2.2 kb and the insert is 131 bp, so for 100 ng of backbone there should be ~20 ng of insert
- Gel extract plasmid to get final library.
Today, we did PCRs and PCR purifications for step one as outlined above. We tested the concentrations of the results, displayed below:
In order to have enough backbone to work with, we seeded some LB/spec with CB bottom bacteria in preparation for 4 minipreps of 3 mL each tomorrow. This should definitely yield the 1800 ng of backbone we will need for the ligation.
Practice plate primer tag test
We tested the primers to see if their selection of subpools was exclusive enough or not (i.e. to make sure that our control primers did not amplify the CB bottom sequences, etc.). We ran a PCR crossing primers with DNA with the following results:
July 30th
Team TolC
- Checked the plate with ECNR2 with Zeocin knocked into the place of rpoZ. There are appear to be some 15-20 colonies on the plate. This strain is ready for transformation with the zif268 plasmid
Team ZF
CB bottom miniprep
Today we performed a miniprep on the 4 tubes of CB bottom bacteria with 4 ml of culture in each tube. 6 ml of culture were eventually consolidated into a single spin tube during the miniprep, so this would be the equivalent of 4 minipreps in one tube. Of note, after lysis (during addition of buffer P2), the tubes turned extremely blue, darker blue than we had ever seen before. This would seem to indicate that the lysis step occurred very successfully. However, even after consolidating four minipreps into one tube, we still got terrible concentration results for our samples, displayed below:
We will thus seed more LB/spec culture with CB bottom bacteria tomorrow, and perform the miniprep on Monday with TF supervision to hopefully troubleshoot some of the problems with low yield.
Also of note is that the bacteria were left on the shaker for approximately 23 hours at the time of miniprep, which is longer than the ideal 12-14 hours.July 31st
Team ZF
Primer tag test two
On Friday, we repeated the PCR to test for sub-pool specificity of our primer oligo tags for the chip, using the same PCR protocol. Today, we ran a gel to see if the results were any different, but alas, they were the same. It seems that the primers for CB bottom also amplify out the control sequences, which means that our primer tags are not as specific as we thought they were. The gel can be seen below, with the lanes arranged as follows: CB primer + CB DNA, control primer + CB DNA, control primer + control DNA, and control primer + CB DNA. We would ideally expect bands in lanes 1 and 3 and no bands in lanes 2 and 4. However, this was not the case:
We might try doing touchdown PCR or raising the annealing temperature to get more specificity in the future. It may also be worth ordering oligos to represent the other 5 subpools of targets and cross test the primers with each DNA subpool to make sure that the primers are specific enough to pull out only one subpool. In the end though, this specificity is not critical to our success, and so it is ok if it doesn't eventually work.
Preparing for another CB bottom miniprep
Today we seeded 4 tubes of culture with 100 ul of CB bottom bacteria at saturation. Each tube carried 4 ml of culture. The tubes were put on the shaker at approximately 6:30 pm, so they will be ready for miniprep tomorrow morning after the lab meeting.August 1st
Team Wolfe
Optimizing PCR reaction for pSB4K5:
The gradient PCR did work, but the yield was still very low. We will try a few different approaches to increase our PCR product. All these reactions were done only with 3:11P (since 2:17H showed no product in the gradient).
1) Gel Stab PCR:
- We ran out more of the gradient PCR product (see image below) and used it as the template for a new PCR
- Gel stab went into 20µL H2O; 66 C annealing, 3 min elongation; pSB4K5_F+ZFBhl and pSB4K5_R+URA3hl primers
- Results: unsuccessful. Perhaps the gel stab did not give an adequate amount of template.
2) Reverse Touchdown PCR:
- Because the primer initially anneals with 20 bases and then with 60, we will start with a low annealing temperature and then raise it as the overlapping region grows.
- 58-66 C annealing, changing 1 degree per cycle (9 cycles); the rest of the 25 cycles were done at 66 C
- 3 min elongation
- Results: one of the four reactions worked. This is very strange since the reactions should have been identical.
3) Template concentration gradient PCR:
- The primer dimers may have been forming since there wasn't enough template to compete with the primer-primer interactions.
- All reagents were kept the same except template: 1µL, 1.5, 2, 2.5, 3, 3.5, or 4µL were used
- 66 C anneal, 3 min elongation
- Results: only one of the reactions worked. We did use the gradient cycler which has not always worked well for us. We also noticed that when we removed the tubes from the cycler, most of the liquid had condensed on the lids.
4) Gradient PCR (overnight):
- Since our original gradient seemed to have been one of the more successful ways, we will repeat it but with a tighter temperature range
- Normal KAPA protocol, annealing 65-69 C, 3 min elongation
ZFB-wp-his3-ura3:
- For isothermal assembly we need a high concentration of each of the DNA components, so we decided to PCR more of our ZFB-wp-his3-ura3 insert and use the Qiagen Minelute kit to purify.
- primers: ZFB-wp-f and ZFB+kan_short_R; ZFB-wp-hisura template
- because of a substantial difference in primer melting temperatures, we did a 57-61 C annealing gradient; 90 sec elongation
- Results: all 8 conditions worked!
- Minelute PCR purification of 4 of the reactions: yield=epic.
pSR01 plate reading:
- We shared a 100-well plate with TolC and will take readings overnight with the following conditions:
- Wells 51, 61...91: LB+tet/amp
- Wells 52, 62...92: NM+his
- Wells 53, 63...93: NM
- Wells 54, 64...94: NM+1mM 3-AT
- Wells 55, 65...95: NM+2.5mM 3-AT
- Wells 56, 66...96: NM+5mM 3-AT
- Wells 57, 67...97: NM+his+100µM IPTG
- Wells 58, 68...98: NM+100µM IPTG
- Wells 59, 69...99: NM+1mM 3-AT+100µM IPTG
- Wells 60, 70...100: NM+5mM 3-AT+100µM IPTG
- 51-60: empty
- 61-70 and 71-80: inoculate with 1µL of selection strain culture
- 81-90 and 91-100: inoculated with 1µL of selection strain + pSR01 culture
Team ZF
Restriction enzyme cut test, take 3
We repeated the restriction enzyme reaction once more to obtain a better and more definitive gel image, and we used the following formulation:
- 8 ul of 1x BSA/Buffer 4
- 2 ul of DNA (used the 79.4 junction PCR product again)
- 0.5 ul of BsaI-HF
This time it was incubated at 37*C for 15 minutes, and the product was PCR purified.
1.5 ul was run on the gel.
Redoing the isothermal assembly/transformation for Pos con 77 and the 3 controls
Today we redid the isothermal assembly for Pos con 77, Zif268, OZ052, and OZ123 using our previous protocol. We also performed a chemical transformation using Top 10 ChemComp cells, and plated them with 100 ul each plate. We will have to wait and see whether colonies grow tomorrow.
We also performed a cross junction PCR on the isothermal assembly product to make sure that it worked, using our usual protocol. The gel (pictured below) shows appropriately sized bands for the cross junction.
Practice plate library assembly
We cut 20 ul of CB bottom plasmid using the following recipe:
- 2 ul of 10x Buffer 4
- 16 ul of DNA (at 40 ng/ul)
- 1 ul of enzyme
- 1 ul of BSA
This was run out on a gel, and the band at size 2756 bp was extracted. This will be extracted and used in a ligation reaction tomorrow.
Team TolC
Lambda red recombination of zeocin into ECNR2
- Performed electroporation and lambda red on ECNR2 in order to insert zeocin for rpoZ knockout, in order to have for control in plate reader
- Performed PCR and ran gel on products
- Successful!!
- Performed PCR and ran gel on products
Transformation of Zif268 into ECNR2+zeocin without Kan+ZFB+wp
- Performed transformation of Zif268 on spec plasmid into ECNR@ without Kan insertion in order to have a positive control against our selection system
- Run PCR on product tomorrow to determine success
Observation of growth through plate reader
- Divided plate up in to 45 different cultures
- 1-15 contained ECNR2+kan-ZFB-wp+zeocin+Zif268 spec plasmid
- This is the selection system we created
- They were grown in .005% SDS and 2,4,8,12,16 µM of IPTG
- 16-30 contained ECNR2+Kan-ZFB-wp with out Zif268 spec plasmid
- This served as a negative control
- Also grow in .005% SDS and 2,4,8,12,16 µM of IPTG
- 31-45 contained solely ECNR2
- This served as a postive control
- Grown in .005% SDS and 2,4,8,12,16 µM of IPTG
- 1-15 contained ECNR2+kan-ZFB-wp+zeocin+Zif268 spec plasmid
- Plate was taken down to plate reader at 5:25 pm and began a 16 hour program
August 2nd
Team TolC
- PCR to check for spec plasmid in ECNR2+zeocin strain successful. The cultures are taking a long time to grow, though, so we might make another transformation with the zif268 plasmid tomorrow, if they do not appear to have grown by tomorrow
- IPTG appears to be not working properly. It appears to retard growth. It is possible that the IPTG here may not be working, so an aliquot of IPTG from the Wyss should be brought in tomorrow.
Team ZF
Checking on the Pos Con 77 and F2/F3 Control Plates
All our plates grew bacteria colonies. At the end of the day, we picked 4 colonies from each plate, and grew them in 2 mL of LB+spec.
Miniprep of CB Bottom for ligation
We performed another miniprep on the CB Bottom plasmid, but this time we concurrently performed a miniprep on PZE22G that had been grown overnight in LB+Kan.
Because the yield of the PZE22G miniprep is in the range that we expect (100-200 ng/ul), while the yield of the CB Bottom miniprep is still so low, we concluded that our problem is likely due to the plasmid or LB+spec media. We made new LB+spec. In the future, we could also pick new colonies and grow them up in cultures.
We were planning on using the miniprep product in our ligation reaction. We originally thought that we would need 100 ng of DNA for the reaction, but then we were told that we could have as little as 20 ng of DNA. We decided that in that case, our current yields were sufficient.
Ligation of CB Bottom backbone and F1
We wanted to insert an F1 into a CB backbone today. Yesterday, we cut the CB Bottom plasmid, in preparation for the ligation reaction. Today we gel purified the product. It was difficult to tell exactly which band was the one we were looking for, so we cut out 2 just to be safe. Our gel purification yields were:
- Top band: 23 ng/ul
- Bottom band: 16 ng/ul
We attempted to cut the F1 oligo from the plate using Bsa1. Our product after cutting was 79 bp, which is technically smaller than the 100 bp minimum for PCR purification. However, we attempted the PCR purification anyway, because this cut-off is unlikely to be strict. We then ran a gel to see the band size of the product that remained. Because our first gel was unsatisfactory, we repeated the digestion a second time.
Attempt One:
- Digestion reaction recipe:
- 2ul DNA
- 8 ul 1x BSA/Buffer 4 solution
- 0.5 ul Bsa1
- PCR purification yield: 34 ng/ul
Attempt Two:
- Digestion reaction recipe:
- 2ul DNA
- 1 ul Bsa1
- 50 ul buffer BSA
- PCR purification yield: 33.5 ng/ul
- We also kept a sample of unpurified PCR product from this digestion to run on a gel, to see if we're losing our desired product in the PCR purification step.
We performed a ligation reaction using the following recipe:
- 5 ul ligation mix
- 4 ul backbone
- 1 ul oligo
But after seeing the gel results we are unsure whether the ligation could be successful.
In order to further troubleshoot our less-than-ideal gels for our digestion product, we ran a third digestion reaction, using the second recipe. This time we simultaneously digested our oligo, and our cross junction (which had been shown to work before). We also ran the unpurified PCR product from recipe 2 on this gel.
See tomorrow's entry for the gel image. Our digestion of the cross junction appeared to have worked, while once again, we got unusually large bands for our oligo.
PCR of Zif268 to create GFP Reporter Strain
We performed a PCR to clone out Zif268 so that we can later assemble Zif and GFP into our reporter plasmid. We used the Zif clone Forward and Zif clone Reverse primers (Primer index: 148 and 149). We had an annealing temperature of 62* and an extension time of 90 seconds. Our expected product was ~3 kb.
See tomorrow's entry for the gel. Our PCR appeared to have worked, and we have a strong band at our expected product size.
Team Wolfe
Plate reader:
- We got some beautiful growth curves! A few of the more critical ones are below:
- NM+his: both selection strain and pSR01 grew, but the selection strain grew faster. This might just be an artifact from having accidentally added more cells to the selection strain than the pSR01 wells, but it may also show that the pSR01 strain grows slower. Another possibility is that we are expressing enough zinc fingers for it to be slightly toxic for the cells. Next time maybe we should let the plate reader go for longer than 16 hours just in case the cells need more time.
- NM: selection strain does not grow, while pSR01 does a perfect rescue!
- pSR01 HARV3-AT comparison: the concentration of 3-AT does not seem to make a difference in the growth curves (the differences appear to be due to different wells starting with slightly different amounts of cells). Next time we can try a higher 3-AT concentration, but since we expect Zif268 to bind strongly this might just be what you get.
- NM+his+IPTG: IPTG more or less killed the pSR01 cells but not the selection strain. This may be caused by too many zinc fingers being expressed, so we may want to try less IPTG.
EcNR2 Wolfe Selection system: MAGE
- Our MAGE oligos are finally here! We performed 3 rounds of MAGE and designed oligos for allele-specific PCR to do tomorrow to check our colonies.
- 2µM final concentration HisB VE deletion and PyrF KO oligos; 2.5-3hr recovery between rounds; plated 1µL, 10µL, and 100µL
pSB4K5:
- Our pSB4K5 PCR still eludes us. Our overnight gradient did work, but no better than the other days' reactions.
- Gel extraction and Minelute only gave us a 9.4ng/µL yield (260/280=1.82) which is not enough for isothermal assembly. We will have to miniprep more plasmid for template for PCRs, and we might want to consider using ligation or some other method.
August 3rd
Team ZF
Yesterday's last digestion and PCR results
We ran a gel of our digestion product from yesterday, and our PCR to clone out Zif 268 to construct the reporter plasmid.
Innoculation of Zif 268
We were running low on Zif 268 plasmids, so we decided to make a new culture from the glycerol stock. It has been left in the 37* shaker overnight.
Sending Pos Con 77 and swap controls for sequencing
We performed a miniprep on our cultures of Pos Con 77, and OZ052 swap, OZ123 swap, and Zif 268 swap.
Once again, we got unusually low yields. We decided to test the quality of our spec, to see if that was the problem. We grew PZE22G(which does not have spec resistance) in spec to see if it grew. By the end of the day, nothing had grown.
Still, we decided to PCR the miniprep product, since any DNA that is in the product should be amplified. We performed our usual protocol for a cross junction PCR, but we repeated the reaction for 30 cycles.
Some of the bands varied in size, but we decided to send it all in for sequencing.
Constructing the reporter Zif268 Plasmid
PCRs of Zif 268 and GFP
We decided to perform more PCRs to clone out Zif 268, since it will have to be gel purified. This reaction had the same conditions as yesterday's.
We also received the GFP plate today, so we decided to perform a colony PCR in order to get out the sequence that encodes GFP. We used our GFP Forward and GFP Reverse (Primer Index: 146 and 147) for the reaction, had an annealing temperature of 58*, and an extension time of 30 seconds. The expected product size was ~700 bp.
Gel Purification of Zif 268 and GFP
We performed a gel purification of our PCR products.
After gel purification, Zif 268 had a concentration of approximately 110 ng/ul.
While we had a bright band for Zif 268 on our gel, our band for GFP was much fainter, so we decided not to gel purify it, and instead to re-do the PCR (4 times) for that reaction.
Our yield for the gel extraction of GFP was 10.2 ng/ul.
Isothermal assembly and transformation
Our yield for the gel extraction was lower than what we wanted, but we figured we would try the assembly anyway. This meant that we needed 2.9 ul GFP and 0.9 ul backbone. Ideally all the DNA takes up 2.5 ul when performing an isothermal assembly. We decided to perform 2 reactions, so that one of them might work.
- First recipe (total volume 10 ul):
- 1.9 ul GFP
- 0.6 ul backbone
- 7.5 ul isothermal assembly mix
- Second recipe (total volume 11.3 ul):
- 2.9 ul GFP
- 0.9 ul backbone
- 7.5 ul isothermal assembly mix
We performed a transformation using ChemComp cells, plated 100 ul, and left them in the incubator overnight.
Practice plate ligation
After seeing the results of our third oligo digestion yesterday, we were confused about why the reaction was not working. After talking with our teammates, we realized that we had been performing the reaction on single stranded DNA, instead of our double stranded PCR product. We performed the digestion again today with oligos 1 and 2, using the following recipe:
- 15 ul DNA
- 0.5 ul enzyme
- 85 ul buffer/BSA
After letting the reaction sit at 37* for about 45 minutes, we performed a PCR purification, and then ran the product on a gel:
Finally, we had gotten bands at the appropriate size! We noticed that there were multiple bands smaller than our uncut oligo. We figured the smallest band was the one we were looking for. The one that was slightly larger was likely the result of the enzyme only cutting at one end of our oligo. We wondered whether all of our product would have been the appropriate size if we had let the reaction sit at 37* for a longer period of time. Regardless, we decided to use this product in our ligation reaction. If both ends of the oligo aren't cut, then it should not be able to bind with the backbone.
We performed a ligation reaction with our cut CB Bottom and our digested oligos, using the following recipe:
- 5 ul ligation mix
- 4 ul backbone (~20 ng/ul)
- 1 ul insert (~15 ng/ul)
This would leave to a vector to insert ratio of 1 to 8, which is what is normally used for ligation reactions.
This is how we calculated the amounts of inserted needed for our ligation:
PCR product: ~60 ng/ul We want 1 ug of DNA to be cut, so that after the PCR purification, when we elute in 30 ul we will get a concentration of ~30 ng/ul.
In practice, we end up losing some of our product, so we ended up with 15 ng/ul.
After letting the reaction sit at room temperature for 1 hour, we transformed ChemComp bacteria with the ligation products. We plated 50 ul and 400 ul of the transformation and left the plates at 37* overnight.
Team Wolfe
- Overnight MAGE round 3 colonies grew - 1µL, 10µL, and 100µL used with (too)many cells on 100µL plate and a workable amount on the 1µL plate.
- MAGE round 3 PCR
- 10µL reactions
- Primers: HisB VE KO Check_R, His_F and PyrF KO Check_F, PyrF IR
- 47 colonies tested for knock out of HisBUra3 and 47 colonies tested for knock out of pyrF
- HisB Control - selection strain with same primers
- PyrF Control - Colony 1 with PyrF forward and PyrF internal reverse primers
- Reverse Touchdown PCR to obtain the kan backbone with homology from pSB4K5 biobrick plasmid:
- Used annealing temperature range of 58 to 68 with two rounds elongation at 2 degree intervals and 15 elongation cycles at 68 degrees.
- PCR failed
- Miniprep of pSB4K5 strain for future PCR yielded 15ng/µL
- Plate reader
- Wells 1, 8, 15... 36 LB+tet/amp
- Wells 2,9,16...37 1mM 3-AT
- Wells 3,10, 17...38 2.5mM 3-AT
- Wells 4,11, 18...39 5mM 3-AT
- Wells 5,12, 19...40 10mM 3-AT
- Wells 6,13,20...41 25mM 3-AT
- Wells 7,14,21...42 50mM 3-AT
- Wells 1-7 = empty
- Wells 8-14 = selection strain
- Wells 15-28 = single selection strain + pSR01 colony (duplicates)
- Wells 29-42 = second selection strain + pSR01 colony (wells 33-35 were not usable)
- MAGE rounds 4&5 completed, colonies plated at 1µL, 10µL, and 100µL volumes.
Team TolC
- Set up and ran plate reader for the growth rates of ECNR2+kan-ZFB-wp+zeocin+spec plasmid, ECNR2+kan-ZFB-wp+zeocin Δspec plasmid, ECNR2, and ECNR2+zeocin+spec plasmid Δkan-ZFB-wp in .04% SDS and 25, 12.5, 6.25, 3.125, 1.5625 µM
August 4th
Team ZF
Pos Con 77 and control swap sequencing results
| Pos. Con. 77 | Notes | |
| 1 | lots of snps and deletions | |
| ✓ | 2 | G->C |
| 3 | deletion before type II cut site | |
| 4 | deletion in terminator | |
| OZ052 swap | Notes | |
| ✓ | 1 | G->C |
| ✓ | 2 | G->C |
| 3 | deletions and mismatches in ultramer | |
| 4 | terrible | |
| OZ123 swap | Notes | |
| ✓ | 1 | fine, deletion at end |
| 2 | deletions and snps | |
| 3 | long deletion in helix | |
| ✓ | 4 | fine |
| Zif268 swap | Notes | |
| ✓ | 1 | fine, TGEKP instead of TGQKP |
| ✓ | 2 | G->C, TGEKP instead of TGQKP |
| 3 | terrible | |
| ✓ | 4 | G->C, TGEKP instead of TGQKP |
We had a good sequence for all of our samples. We made 2 ml cultures of these, to miniprep, make final plates, and glycerol stocks. Note, that for Zif 268, the linker between F1/F2 is 'TGEKP', as that was the sequence in the ultramer we ordered. The actual sequence is 'TGQKP'. If this proves to be a problem, we can always try using quick change to insert the appropriate codon.
Checking on the reporter strain
Our transformed bacteria with the 11.3 ul isothermal assembly product and 100 ng backbone grew. We picked four from the plate and are growing 2 ml cultures from time to miniprep and send for sequencing tomorrow.
Our other bacteria with 10 ul product, but less than 100 ng backbone, did not grow.
Ligation of oligos and backbone
Checking our plates
No colonies grew on any of our plates that contained bacteria transformed with ligation product. We wondered if this was due to our ligase no longer working, so we decided to test our ligase.
Testing our ligase
We wanted to test our ligase by cutting our cross junction with Xba1, and then ligating it, so see if it formed the original cross junction. We ran a gel comparing our whole cross junction, our digested product, and our ligated product.
Our uncut cross junction was ~1.4 kb. We saw the appropriate sized bands of 593 and 739 bp after cutting the junction with Xba1. However, after ligation, we saw bands of varying lengths, at the one we expected (at ~1.4 kb) was not noticeably brighter than the other bands. We are unsure what these bands are.
Cutting and gel purifying CB Bottom
We also wondered if our unsuccessful ligation was due to an ineffective CB Bottom segment. We had been unsure exactly which band was the appropriate one, and so we cut both. Just in case we decided to re-do the digestion of the CB Bottom plasmid, with Bsa1. We used the following recipe:
- 46 ul DNA (to get 1 ug)
- 1 ul enzyme
- 5 ul Buffer T4
- 0.6 ul BSA
We then ran the product on a 0.7% agarose gel:
We cut out the appropriate band, and put it in the fridge to gel purify tomorrow.
Making more Zif268 (whole) plasmid
We performed a miniprep of the Zif268 culture we grew yesterday. The miniprep had a yield of ~16 ng/ul.
Team Wolfe
Plate Reader
- Looks great!
- 1mM, 2.5mM, and 5mM 3-AT have slight differences in growth rate, but it is hard to tell how significant they are. However, starting with 10mM 3-AT, the growth rate drops dramatically! Here is an example from 1 culture set:
MAGE 3 HisB and PyrF Knock Out Gel
- HisB Check
- Control reaction showed a nice strong band. All the other lanes had bands, but they varied in strength. It looks like we may have some hits, but we need our WT control primers to come in for us to be sure.
- PyrF check
- Similar to HisB--all the lanes had bands but some were brighter than others.
ZFB-wp-his3-ura3 restriction enzyme PCR
- PCR to add Spe1 and Xba1 sites to join insert with pSB4K5 plasmid
- Spe1 ZFB_F and Xba1 URA3-R primers
- 90 sec elongation, 62˚C annealing
Team TolC
Plate Reader
PCR of plate reader colonies
- Made glycerol stock of ECNR2+spec plasmid+zeocin, WS
August 5th
Team Wolfe
Spe1 ZFB-wp-his3-ura3 Xba1 PCR purification:
- Minelute: 595 ng/uL, 260/280=1.87
pSB4K5 miniprep:
- To get more pSB4K5 3:11P we did a miniprep of a 6 mL culture
- Yield: 8ng/uL
- This yield is still very low, but a gel showed that DNA is present in the sample.
File
ZFB hisura digest:
- 40uL total volume:
- 4uL buffer 4(10x)
- 0.4uL BSA (100x)
- 2uL Spe1
- 2uL Xba1
- 2uL Spe ZFB-his3-ura3 Xba1 (about 1ug)
- 29.6uL H2O
- incubate overnight at 37 C
Prep for tomorrow:
- pSB4K5 3:11P culture: repeat miniprep tomorrow and hope for a better yield
- selection strain + pKD46: to transform with finished ligation product
- selection strain: streak new plate
Team ZF
Gel extraction of cut CB bottom plasmid
We performed a gel extraction on the CB bottom plasmid that we cut with Bsa-HF and ran through a gel yesterday. However, we got an extremely low yield of 3.4 ng/ul.
After talking with Dan about the not-so-ideal experiences with Bsa-HF, we decided to make a few changes to our protocols, and that we would try the following to see how they would work:
To troubleshoot the poor oligo cutting:
1.5 ul of enzyme (20 units/ul)
2 ul of NEB buffer 4
0.5 ul of BsaI-HF
16 ul of DNA
20 ul total
Incubute for 1.5 hour in a thermal cycler at 37 degrees.
Denature at 65 degrees for 20 minutes and then pcr purify.
For troubleshooting the poor plasmid cutting:
Do the same thing but do it for longer (2 hours) and also run an uncut plasmid to see which bands come up.
We checked to see if dcm methylation happens for bsaI at our cut sites, but there were no signs of the dcm recognition sequences (CCAGG or CCTGG) anywhere near them.
GFP reporter plasmid sequencing
Today we miniprepped the four GFP colonies that we picked yesterday in order to run a junction PCR on them and send them out for sequencing. Our junction PCR used the previous protocol's annealing temperature of 55 degrees, but the extension time was lengthened to 1:15 to account for the extra length of the GFP insert (some additional 700-800 bp). After the PCR, the samples were sent out for sequencing with both the forward junction primer and the reverse 100 bp primer (ZF116 by the primer index).
Glycerol stocks and final platings
We made glycerol stocks of the final four plasmids in Top 10 ChemComp bacteria, which were Pos Con 77 (85.2), OZ052 (52.1), OZ123 (123.4), and Zif268 (Zif268.1). We also made final plates of these bacteria, using 1/1000 ul on spec plates.August 6th
Team ZF
New miniprep protocol
Today, we tried a new miniprep protocol in which the DNA is precipitated out of solution with EtOH, and no spin columns are used. This is because we suspect that our low yields may be due in some part to DNA being washed out of the column prematurely, or DNA not being washed completely out of the column during the elution step. In getting rid of the column, there is no step during which we may lose DNA.
We are extremely happy to report that after weeks of getting dismal miniprep yields, we have found a new protocol that gives us insanely high yields (compared to before)!!! The protocol is the following:
- Centrifuge 1.5 ml of saturated cells at maximum speed for 1 minute, discard supernatant.
- Resuspend cells in 200 ul of Buffer P1.
- Add 200 ul of Buffer P2 to lyse the cells. Mix by inverting several times, and let sit for 3 minutes.
- Add 200 ul of Buffer N3 to precipitate SDS, mix by inverting several times.
- Centrifuge at 4 degrees for 15 minutes.
- Transfer supernatant to new tube, add 1 ml of 100% EtOH to precipitate DNA.
- Centrifuge for 5 minutes at RT. At this point a DNA pellet should be visible at the bottom of the tube.
- Discard EtOH, wash DNA pellet with 1.5 ml of 70% EtOH (this gets rid of salts).
- Centrifuge for 5 minutes at RT.
- Discard EtOH, let tubes dry for approximately 6-8 minutes by inverting them on a piece of Kimwipe tissue.
- Resuspend DNA pellet in 20 ul of heated EB buffer.
Our resulting yield was overall 463.4 ng/ul!! The purity also looked extremely good, with a 260/280 value of 1.98. The nanodrop results can be viewed below:
We will have to repeat this again tomorrow to see if we get the same yields with this procedure. Also, we should run some of these out on a gel to verify what it looks like.
New restriction enzyme protocol
The protocol used for the restriction enzyme digest will be as described yesterday:
- 16 ul DNA (any concentration)
- 2 ul 10x buffer 4
- 0.5 ul 100x BSA
- 1.5 ul enzyme (30 units)
This gives a 20 ul total reaction volume. The reaction is incubated at 37* for 2 hours, denatured at 65* for 20 minutes, and then left at 4* for storage overnight.
A restriction digest using this protocol was performed on practice plate oligos 5 and 6 (reactions for oligos 3 and 4 were no good due to a pipetting mishap).
Sequencing results for the GFP reporter plasmid
Due to Seqman crashing and the Harvard VPN not being active, we will have to wait until Monday for these results.
Team Wolfe
MAGE round 3 allele-specific PRC:
- did 4 types of PCR: HisB KO (HisB VE KO check_R and His_F), HisB WT (HisB VE WT check_R and His_F), PyrF KO (PyrF KO check_F and PyrF_internal_R), PyrF WT (PyrF WT check_F and PyrF_internal_R)
- 24 colonies: A10, A11, C2, C3, C6, C8, C9, D1, D5-12, E3, E7, F2-4, G2, G3, G9
- 10uL total reaction volume
- same protocol as earlier reaction (64C anneal, 1min elongation, 25 cycles)
- HisB bands at about 900bp; PyrF bands at about 400bp.
- Results: none of the colonies have both genes knocked out but some do look like they may have the HisB deletion. We may try again using MAGE round 5 colonies.
pSB4K5 digestion:
- Since our minipreps have been giving such low yields, we tried an [http://openwetware.org/wiki/Knight:Miniprep_low_copy_plasmids altered protocol]. The result, however was the same: 6mL of culture produced 19ng/uL (in 30uL)
- PCR purified Spe1 Xba1 ZFB-wp-his3-ura3 digestion (Minelute): 53.8ng/uL 260/280=1.82. This is interesting, since it appears that half our DNA has been lost.
- pSB4K5 Spe1 and Xba1 digestion: 72uL total
- about 1mg DNA=60uL
- 2uL each enzyme
- 7.2uL buffer 4
- .72uL BSA
- 3 hour incubation at 37C
- gel extraction: the gel was empty. No clue why. Obviously this is a setback--we will have to redo the miniprep and digestion before we can form the whole plasmid.
August 7th
Team ZF
Miniprep repeat with new protocol
Another miniprep was repeated on 3 ml of CB bottom plasmid culture using the protocol from yesterday, with a yield of 1 ug/ul. This seemed very high considering our previous results, and we are suspicious that there may be some genomic DNA contamination. Contamination may occur either from leaving cultures on the shaker for too long (with cells lysing and releasing their genomic DNA) or from shaking too hard during the lysis process (shearing chromosomal DNA and allowing it to escape the cell). The lysis process was performed by mixing through tube inversion, so this is likely not the cause. The tubes that were miniprepped yesterday were left on the shaker for approximately 22 hours, while the tubes from today were on the shaker for 17 hours. Strangely, although the tubes were on the shaker for a shorter amount of time, there was still higher yield.
The concentration and absorbance curve data can be seen below:
As a note, the tubes were unusually dark blue this round after addition of the lysis buffer (buffer P2).
The uncut plasmid was also run out on a gel alongside the cut oligos (loaded 0.5 ul). This should theoretically have been around 500 ng of DNA, but for some reason it only shows up very faintly. This is not good news.
Results for new restriction enzyme protocol
The resulting reaction from the oligo cut with Bsa-HF yesterday was run out on a gel with the following results:
There appears to be a band of the correct size. However, there is still incomplete cutting as indicated by the second band at around 100 bp, but there is no longer a band at the uncut position, indicating that all of the oligos underwent a single or double cut.
The plasmid DNA from the miniprep today was also put through a restriction digest using the same 2 hour protocol as the oligo digest. The result was then PCR purified, but the yield was extremely low compared to the input. Using 16 ul of DNA, there were 16 ug of plasmid that went into the restriction digest. After PCR purification and elution in 30 ul of EB, the yield was only 28.5 ng/ul, meaning only 0.855 ug of DNA in the purified product. This translate to over 95% of the DNA somehow being lost in the PCR purification. We are at a complete loss as to why this has happened...
The cut plasmid DNA was run alongside uncut plasmid in a gel, with the results below:
August 8th
Team Wolfe
MAGE sequencing
- Colonies
- A10, C3, and D9 from round 3 MAGE
- A1-D8 round 5 MAGE (colonies picked today).
- Primers for PCR
- His_F, His_R
- PyrF_F, PyrF_R
- Primers for sequencing
- His_R, PyrF_F
- 64 degrees annealing and 90 seconds elongation
- Gel shows PCR was successful
5-FOA Concentration Gradient
- Protocol recommended 2.5mM conc. FOA but we tested lower concentrations because FOA = $$$: ($FOA>$Gold)
- 61,68...89 NM+His
- 62,69...90 NM
- 63,70...91 .1mM 5-FOA
- 64,71...92 .25mM 5-FOA
- 65,72...93 .5mM 5-FOA
- 66,73...94 1mM 5-FOA
- 67,74...95 2.5mM 5-FOA
- 61-67 = selection strain colony 1
- 68-74 = selection strain colony 2
- 75-81 = pSR01 colony 1
- 82-88 = pSR01 colony 2
- 89-95 = empty
pSB4K5
- Since minipreps are not working we will try a midiprep
- Innoculated 100mL LB+Kan with a pSB4K5 3:11P colony
Streaked Plates
- Selection strain
- Selection strain + pSR01 plates because they were getting old
Team ZF
GFP Sequencing Results
We were able to access the results of our GFP sequencing.
| GFP Forward | Notes | |
| 1 | 1 deletion at end | |
| ✓ | 2 | perfect |
| 3 | Insert of G | |
| 4 | perfect | |
| GFP Reverse | Notes | |
| 1 | 4 snps | |
| ✓ | 2 | 1 deletion at end |
| 3 | 0 snps | |
| 4 | snps |
However our sequencing failed to cover the the Shine-Dalgarno region. We decided to test our strains by growing them on a 96 well plate. Each well had 150 ul LB+spec. Colonies designated 1-4 are 5 ul of the ones we sent out for sequencing. Those designated 5-8 were 2 ul colonies which were picked from our reporter plate then and diluted in 10 ul LB+spec. The plate had the following configuration:
- Rows A and B, 1 through 8: New colonies picked from the plate of our reporter strain.
- Row E, 1 through 8: Colonies 1-8 in 0 ul IPTG
- Row F, 1 through 8: Colonies 1-8 in 2 ul IPTG
- Row G, 1 through 8: Colonies 1-8 in 10 ul IPTG
- Row H, 1 through 8: Colonies 1-8 in 100 ul IPTG
After shining blue light on the plate, nothing glowed. We realized that this was because we did not have a spacer between the Shine-Dalgarno sequence and the start codon, meaning the ribosome would not be able to reach both sites. After doing some reading, we realized that a spacer of 6-10 bps is ideal. We will design this tomorrow.
Re-do minipreps for control backbones with new protocol
We used the new protocol to do a miniprep on the control backbones.
Our yields were extremely high with the new technique--it was difficult to believe. We were a bit wary of the nanodrop readings so we decided to run the product on a gel and compare the brightness of the band to the ladder in order to determine the actual concentration of DNA present in the miniprep product.
The first six lanes are our miniprep product. The next four contain serial dilutions of the Zif 268 plasmid. We determined that it looks like our miniprep product actually contains ~80 ng/ul of DNA, after comparing it with the 1650 bp band.
Ligation and transformation of Oligo 5 and 6 and CB Bottom
We decided to try the ligation of our oligos and the backbone, using our previous protocol. We then transformed the product using ChemComp cells, plating 50 and 450 ul.
Cutting the Oligo Pool
We decided to try cutting a pool of our 18 oligos. We used our new recipe, adding 0.78 ul of each oligo so that the DNA takes up a total of 16 ul. We followed the new protocol for one reaction, and the other digestion was left to run overnight. We will compare the results tomorrow.
Team TolC
Plate Reader
- Set up and began running 160 well plate reader
- Plate was set up as follows:
PCR on TolC
- Have failed twice before in trying to PCR out the TolC sequence from the WS and WN strains, which are ECNR2 without the Kan-ZFB-wp insert, worried that TolC may have somehow been deleted or removed which seems very unlikely
- Ran third PCR and gel and failed again
- NOTE. It's possible that the ECNR2delTolC strain was used, but that still doesn't explain the 800bp band using the colonies from the zeocin plate used for selecting the zeocin cassette insertions that led to the WN strain (Noah's PCR 8/9/2011). It's strange given that the colonies off that plate give different results for the TolC region flanked by the TolC_seq primers, given that the zeocin PCR shows that all colonies have the zeocin insert (primers are Zeocin_R and rpoZ_R)
August 9th
Team Wolfe
Sequencing check for HisB and PyrF deletions
- One colony definitely has both deletions! It is now being grown for lambda red and glycerol stock
Zeocin Lambda Red
- Made 10 zeocin plates (zeocin concentration=50µg/mL)
- Grew up cells from D2 MAGE#5 to midlog and used 1mL for lambda red
- 2µL zeocin cassette (about 300ng)
- recovered for 2 hours before plating
pSB4K5 Digestion
- Enzymes
- 2µL spe1
- 2µL xba1
- DNA = pSB4K5 plasmid (1µg, 17.2µL)
- 4µL buffer 4
- .4µL BSA (100x)
- 14.4µL H2O
- incubated for 2hrs at 37C
5-FOA concentration gradient results
- Since 5-FOA is more than worth its weight in gold, we were hoping that lower concentrations than recommended in the protocol would work for inhibiting the growth of cells expressing pyrF/URA3. According to last night's plate reader experiment, concentrations as low as .1mM completely stop cell growth!
- Also, another nice growth curve showing pSR01's rescue of the selection strain in incomplete media:
Team Wolfe-ZF Collaboration
Midiprep
- Note: Instead of second lysate clearing spin, supernatant was transferred to falcon tube and poured into column
- Yields
- pSB4K5 = 58ng/µL, 260/280=1.84
- Zif268 = 168.52 ng/ul, 260/280 = 1.84
More data on the concentrations can be seen below:
The bands for the midiprep look similar to those from the miniprep using the new ethanol precipitation protocol, except they are much brighter. In the following gel image, there were 8 ul of plasmid loaded for both the miniprep and midiprep plasmids. It appears that there is much, much less miniprep plasmid than midiprep. Considering that the midiprep plasmid is at approximately 168.52 ng/ul, the miniprep plasmid is probably around our old yield. This means that the Nanodrop reader LIED TO US!! :'( :'( :'(
Here is the gel:
Team ZF
Checking our 5 and 6 Ligation Plate
Colonies grew on all our plates! We ran our cross-junction PCR on 8 colonies each from the 450 ul plates and sent it out for sequencing.
Making more practice plate oligos
We were running low on oligos 1 through 6 from our practice plate, so we did a PCR to make more, using the same protocol as before. We then PCR purified the product.
Design a new primer for GFP
We designed a new primer for GFP, with a 7 bp spacer between the Shine-Dalgarno sequence and the start codon for GFP. According to what we discovered online, it seems that the spacing between the ribosome binding site and the ATG start codon is very important for transcription efficiency. We had previously thought that there were 8 bp from the beginning of the RBS to the ATG codon, but instead it is really 5-9 bp in between the end of the RBS and the ATG codon that is correct. We thus added the spacer that comes directly from the Ambion website, which is AGGAGGTATATACATG (spacer bolded). More info can be found here: http://www.ambion.com/techlib/basics/translation/index.html.
Practice plate oligo pool
This morning, we took out our digestion reactions from the thermocycler, containing all 18 practice plate target oligos mixed together. The tube containing the overnight reaction was a bit crushed by the machine, and it looks like we lost a bit of volume.
Regardless, we performed a PCR purification on the product, and got a concentration of ~17 ng/ul.
Then, we performed a ligation with the Oligo Pool and the CB Bottom backbone using the typical protocol. We performed a chemical transformation with the ligation product, plating at two densities using 50 ul of transformed bacteria and 450 ul. We shall see tomorrow whether any colonies grew.
Yogurt Plate
Yesterday we plated 10 ul of [http://www.stonyfield.com/products/stonyfield/smoothies/10oz-smoothies/peach-0 Stonyfield Super Smoothie] from lunch, which contains [http://www.stonyfield.com/products/stonyfield/what-makes-our-products-so-special/six-live-cultures six varieties of active yogurt bacteria]. Today it grew colonies. It smelled like yogurt this morning. This evening it smelled like rotting yogurt. Yuck.
(UPDATE: According to Dan it smells like Swiss cheese. Now it smells slightly better).
Prepping for western blot tomorrow
We inoculated cultures (4 ml each) in preparation for western blotting tomorrow to verify that the zinc fingers are indeed being expressed by IPTG. Here is the setup:
- Kan - pze22g with GFP as a complete negative control (there should be no zinc fingers in there... hoping there's also no proteins with flag tags in there)
- Spec - Zif268 whole with 0 uM IPTG
- Spec - Zif268 whole with 2 uM IPTG
- Spec - Zif268 whole with 10 uM IPTG
- Spec - Zif268 whole with 100 uM IPTG
We expect to see zinc finger expression to varying degrees in the cultures with IPTG, depending on the level of IPTG. Hopefully we'll have enough protein and FLAG tag antibody to work with tomorrow.
Team TolC
Plate Reader
- There appears to be a significant difference between the KN and KS strands, but only at high SDS levels i.e. 2% (lag of about 5 hours). At lower levels, there appears to be no significant lag, but a difference in the slope of the log phase.
- Even at 0 IPTG, there is expression of the ZF plasmid, suggesting leakiness (constitutively 'ON' promoter for the ZFP)
- WS and WN strands show lack of growth in the presence of SDS at any concentration (except 0) and PCR shows odd results (no band to 800bp bands for the product using tolC_seq_F and tolC_seq_R) add images here
- need to discard the WS and WN glycerol stocks into a 'bad glycerol stocks' box
- need to move the rpoZ MAGE oligos into the bad primers box
OZ052 and OZ123 MAGE into ECNR2+Kan-ZFB-wp+zeocin without plasmid
- Ran two rounds of MAGE for both OZ052 and OZ123 in the KN strain order to create the other two positive controls.
- time constants were 5.2s for the first round, and 5.6s for the second round. After the second round of MAGE, the cells appear to have been growing rather slowly. MAGE 2 was done at 2pm, and the cultures are barely cloudy at 6:28pm
- 3rd round at 10:10pm, time constants are 5.4s and 4.6s
Lambda Red Recombination of zeocin into new ECNR2 culture for rpoZ knockout
- put the colony in 2ml of LB+amp to grow at 3pm. It should be ready for lambda red by 7pm, hopefully?
August 10
Team TolC
MAGE OZ052 and OZ123
- MAGE round 4a, time constants of 5.6s (OZ052) and 1.2s and 5.4s (OZ123). There appeared to be a spark for the OZ123 electroporation (Sarah)
Recombination of Zeocin into ECNR2
- Zeocin insert colonies appear to be growing extremely slowly, only 3 colonies after 14 hours, but may be due to rpoZ knockout
- Growing up culture in case we need to do lambda red of zeocin again
- PCR of the TolC, rpoZ and zeocin bands for colonies 1 and 2 off the zeocin plate.
- numbered, from 1-6, 1-3 is colony 1, 4-6 is colony 2. 1,4=TT 2,5=RR, 3,6=ZR
- PCR successful!! We have the real WN strain now. It should be ready for transformation tomorrow with the zif268 spec plasmid
Team Wolfe
Lambda Red RpoZ Knockout Plates
- Small colonies grew after about 14 hours on zeocin plates
Rpoz Knockout Check PCR
- Picked 24 colonies and used zeocin binding primers as a control and rpoZ binding primers to check for a knockout.
- 48 10µL reactions in total (24 colonies X 2 different primer sets mentioned above)
- Primers: rpoZ_R with Zeocin_R and rpoZ_R with rpoZ_F
- 64 degree annealing temperature
- 90 seconds elongation
Results
- rpoZ was knocked out!
- Reinoculated LB+zeo with culture 9 to use tomorrow for glycerol stocks and transformations
Digestion of pSB4K5 Plasmid
- Gel with yesterday's digestion product suggests that neither of the two restriction enzymes (Xba1 or Spe1) cut our plasmid. This was determined by comparing the band sizes to those obtained by midiporep of the pSB4K5 (whole) plasmid.
- Plan of action: We will use smaller reactions to increase the relative concentration of enzymes and we will use slightly more BSA buffer.
Today's Digestion of pSB4K5 Plasmid
- Xba1
- 16µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Xba1
- Spe1
- 16µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Spe1
- Xba1+Spe1
- 14.5µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Xba1
- 1.5µL Spe1
- Results: the size of the bands isn't what we expected (backbone should be ~3.4kb, insert ~1.2), but the comparison between the different digestions and the uncut plasmid does imply that the digestion we worked. We will go ahead and gel extract from the Spe+Xba digest and use both ligation and sequencing to be sure that this is indeed the plasmid we want.
Team ZF
Mini library ligation results
We checked our plates from our practice plate library assembly yesterday, and we got colonies!! Strangely, our 450 ul plate didn't have a huge number more colonies than the 50 ul plate. Dan suggested that this was because we plated all the cells in a 450 ul volume, which may have sloshed around and taken cells with it. Next time we should pellet the cells and resuspend in 100 ul and plate that. Based on our colony numbers with 50 ul, we should probably plate with 100 ul in the future.
Due to the proliferation of colonies, we picked 90 colonies with the assistance of Will. These 90 colonies should hopefully represent all 18 oligos in equal amounts. We did colony PCRs on all 90 with the junction PCR protocol, and sent them out for sequencing today. We saved these 90 colonies by adding LB/spec to each of the wells after suspending them in the 10 ul of water for the colony PCR. We will eventually harvest and gather all 18 assembled plasmids from this step.
Sequencing results for oligos 5 and 6
Sequencing indicated that most of our colonies worked, and that our F1s were inserted correctly! This is a huge step forward, and gives us good hopes that our 90 colonies also have their F1s inserted. Out of the 8 colonies each that we tested for oligo 5 and 6, only 5.8 (missing chunk on promoter), 5.3 (deletion on linker), and 6.8 (bad trace file) were bad. However, all the rest were good!
We also now have an idea of our combined ligation/transformation efficiency. NUMBERS HERE PLZ
Western blot to detect Zif268
Today we began a western blot in an attempt to detect Zif268 expression from IPTG induction. Due to the fact that the WB gels only have 10 lanes, we dropped the 2 uM IPTG sample for now, keeping the negative control (pze22g + gfp), 0 uM IPTG, 10 uM IPTG, and 100 uM IPTG.
Lysis buffer had the following composition:
- 150 mM NaCl
- 50 mM Tris-HCl
- 5 mM EDTA
- 1% Triton X-100
- 1x proteinase inhibitor
We used the following protocol for lysis:
- Spin down 3 ml of cells into a pellet, discarding the LB supernatant.
- Resuspend cells in 1.5 ml of ice cold lysis buffer, take 200 ul of this into a new tube and add 800 ul of cold lysis buffer.
- Sonicate at power level 2 in two sets of five bursts (each about one second long). Be sure to wait 10 seconds in between sets and keep tube on ice to prevent overheating.
- Spin down at maximum speed for 5 minutes at 4 degrees.
- Take supernatant into a new tube. This is the final protein lysis mixture.
Note! In the future, we should only spin down 200 ul of cells for lysis. According to Alain, most of the time they even just use 100 ul and directly boil it with SDS. May try this protocol tomorrow if current protocol doesn't work (the downside is that we have to start again from the culture stage and not the protein stage).
Prepping samples for the gel:
- For loading duplicates, load 9 ul of 5x sample buffer with 36 ul of protein for a total of 45 ul.
- Parafilm tubes to prevent popping, and boil for 5 minutes at 100 degrees.
- Load 10 ul of protein ladder into gel, and load 15 ul of sample into each well.
Note! For this particular gel, the wells go in this order from left to right: Marker, neg con, 0 uM, 10 uM, 100 uM, neg con, 0 uM, 10 uM, 100 uM, Marker.
Running samples on gel:
- Gel was run at 150 V until the marker band reached approximately 80% of the way to the bottom of the gel.
Transfer:
- Semi-dry transfer was accomplished by wetting blotting paper and nitrocellulose membrane in transfer buffer and making the sandwich on the transfer apparatus. From bottom to top (red to black electrode), the loading order was as follows:
- 2 sheets of blotting paper
- Nitrocellulose membrane
- Gel
- 2 sheets of blotting paper
- Transfer was run at 20 V for 1 hour.
Note! The side containing the protein is face up when the notch in the membrane is situated on the top right hand corner. However, due to troubles with the gel, the lanes are now in the opposite order of loading. So, with the protein right side up, the lanes go: marker, 100 uM, 10 uM, 0 uM, neg con, etc. Also, we forgot to roll bubbles out from the sandwich with the pipette fragment before transferring (my bad!) Hopefully this doesn't impact anything. One last note on the transfer: the current cap was 400 mA, and when we started the transfer, it initially began at 17 V and 400 mA. Over a few minutes, the voltage increased to 20 V and the current began to decrease, hitting 110 mA after a half hour of gradual decay, which is where we expected the current to be.
The transfer seemed to have worked, transferring the marker bands from the gel onto the membrane.
Blocking:
- Membrane was blocked at 4 degrees overnight.
Tomorrow we will proceed with primary and secondary antibody staining and band visualization.
Library ligation optimization
Dan suggests that we optimize the vector:insert ratio for our library assembly. We will try using the following insert ratios (assuming a vector unit of 1): 1, 2, 3, 4, 8, 10, 16. Unfortunately, today we did not have enough cut CB bottom plasmid to immediately begin the ligation and transformation, so we attempted to cut more CB bottom. After performing a cut with Bsa-HF using the previous protocol, we performed a PCR purification on the product, only to find that there were only 3.4 ng/ul of cut plasmid present. This is not enough to perform the ligation, and so we will have to try the CB bottom plasmid cut again tomorrow morning when the cultures have grown enough. We can then do a ligation and transformation tomorrow and count colonies on Friday.
As stated in the earlier section, we should probably plate 100 ul of transformed bacteria to standardize everything and to keep colony counting fairly manageable.August 11th
Team Wolfe
Ligation of the HisUra Insert
- follow [http://www.neb.com/nebecomm/products/protocol2.asp New England Biolabs protocol]:
- 1.5µL of cut Spe ZFB-wp-hisura Xba insert (75ng,a 3-fold molar excess to backbone)
- 6µL cut pSB4K5 backbone (50ng)
- 1.5µL Water (bring volume up to 10µL)
- 1µL Quick T4 DNA Ligase
- 10µL 2X quick ligation buffer and mix
- spin briefly and incubate for 5 min at room temperature
- keep at -20˚C or on ice until transformation
- Transformation: ligation mixture cannot be directly electroporated, so we will start with top10 chem comp cells (see protocols page)
- Following [http://www.neb.com/nebecomm/products/protocol3.asp New England Biolab's transformation recommendations], we used 2µL of ligation mixture in one tube of 50µL cells, and 1µL of unligated cut backbone (about an equal mass) in another tube of cells as a control.
Transformation of pSB4K5 plasmid with His3Ura3 Insert into Chemically Competent Cells
- Transformed into delta HisB delta pyrF delta rpoZ Wolfe strain
- Cells plated in 1mL and 2mL quantities on Kan plates after 3 hours recovery
Transformation of Zif268 Plasmid
- Transformed into HisB delta pyrF delta rpoZ EcNR2 strain
- Cells were plated after 3.5 hours recovery on Spec plates at 10µL and 100µL
Transformation of the pSB4K5 Plasmid with His3Ura3 Insert into Electrocompetent Cells
- Transformed into delta HisB delta pyrF delta rpoZ Wolfe strain
- 3µL DNA from ligation reaction (unpurified)
Team ZF
Western Blot
PCR to get out GFP
Our new GFP forward primer arrived today, so we performed a PCR to get out the GFP region. Because the product will be used for gel extraction, we ran 4 reactions. We had an annealing temperature of ... and an extenstion time of 30 seconds.
Check oligo pool sequencing results
Unfortunately, most of the samples we sent in for sequencing had problems. For many, the primers did not anneal; for others there were poor quality reads. 13 samples in total had good quality reads. Among those we got a pretty good distribution among our 18 oligos. This is promising. We will perform a PCR on 2 random samples to ensure that the cross junction is present. If we get positive results, we will send in 24 more samples for sequencing.
Summary of sequencing results:
| Oligo | Frequency |
| 5 | 3 |
| 6 | 1 |
| 8 | 1 |
| 9 | 1 |
| 10 | 1 |
| 11 | 2 |
| 12 | 1 |
| 17 | 1 |
| 18 | 2 |
Finding the optimal backbone:insert for ligation
We wanted to find the optimal backbone to insert ratio for ligation. Previously we had successfully used 100 ng of backbone, and a ratio of 1 to 8. We decided to try the following ratios:
- 1:1, 1:2, 1:3, 1:4, 1:8, 1:10, 1:16
We ran out of cut CB Bottom backbone, so we needed to digest some more. We performed a miniprep on cultures that grew up overnight, using min-elute columns. Our final concentration of CB Bottom Plasmid was 50 ng/ul and 54 ng/ul.
We followed our usual protocol for digestion, and then performed a PCR purification, once again using a min-elute column to maximize our yield. However, the resulting product only had a concentration of ~5 ng/ul and a purity of ~1.6. We got similarly low yields yesterday, and are unsure where things are going wrong, since we have performed the same procedure in the past with success. Even if the plasmids weren't cutting, they should be present in the PCR purification product, so the low yields are puzzling. We decided to try a midi-prep tomorrow, so see if having more CB Bottom plasmids would help.
We grew up the following 100 mL cultures for midi-prep tomorrow: CB Bottom, Zif268.
Team TolC
Search for a better weak promoter
- We have searched for a better weak promoter than the one we have now and compared their sequences to the present weak promoter in the kan-ZFB-wp construct, sequences were not similar enough to change the weak promoter through MAGE
- To change the promoter would be a big process, so we are thinking of using a negative selection of TolC using colicin and then creating a higher TolC production standard to begin with which would allow the KS and KN growth curves separate more
Transformation of Zif268 Plasmid
- Performed transformation of the Zif268 plasmid into the good ECNR2+zeocin that definitely does have TolC
- Plated 1 mL of culture onto spec plate
Plater Reader
- Kept the IPTG concentration constant at 100 µM while changing the SDS concentration from 2% to 2.5%, 3%, 3.5%, and 4%
- Making two wells per different culture
- Cultures are changing by SDS and IPTG concentration, while also changing from KS, KN, WS, WN colonies
- Since colonies are not near mid-log we may have to fill plate reader tomorrow and run then as well
Gradient Plates
- Created 5 50 mL total gradient plates with 2x Kan in the wedge layer, 20ul of Bluejuice in each 25ml of LB+agar+Kan to mark it with a dye.
August 12th
Team Wolfe
Transformations
- EcNR2 strain with transformed zif268 plasmid grew on spec plates: After 15 hours, 1mL and 2mL plates have ~20 colonies each
- Wolfe strain with transformed ligated pSB4K5 plasmid had no growth on kan plate at 1mL and unusual looking growth on a tet plate at 1mL after 12 hours
- The pSB4K5 plasmid was successfully transformed into the chemically coMpetent cells - will midiprep these to obtain a clean sample of ligated pSB4K5 for another transformation in electrocompetent Wolfe strain cells.
PCR check for Zif268 plasmid insert
- Looks like the plasmid's presence is confirmed by PCR in 8 colonies
- Team ZF's "CJUN" protocol: pZE23G 2123R and pZE23G 3581F primers
- Note well 5 is fainter but is probably due to a loading error where a small volume of sample was left out.
Selection strain genotype confirmation
- We will do one last sequencing round of our EcNR2 selection strain to confirm that HisB and PyrF are indeed knocked out.
- Same parameters as previously described; His_R and PyrF_F used for sequencing
- PCR was successful, as shown by an E gel of several of the reactions.
EcNR2 selection strain plate reader experiment
- We want to see how the EcNR2 selection strain works with and without plasmid, especially in the presence of various concentrations of 3-AT and 5-FOA.
- Wells 1, 14, 27, 40, 53: LB+kan/spec
- Wells 2, 15, 28, 41, 54: NM+his
- Wells 3, 16, 29, 42, 55: NM
- Wells 4, 17, 30, 43, 56: NM+1mM 3-AT
- Wells 5, 18, 31, 44, 57: NM+5mM 3-AT
- Wells 6, 19, 32, 45, 58: NM+10mM 3-AT
- Wells 7, 20, 33, 46, 59: NM+25mM 3-AT
- Wells 8, 21, 34, 47, 60: NM+50mM 3-AT
- Wells 9, 22, 35, 48, 61: NM+0.1mM 5-FOA
- Wells 10, 23, 36, 49, 62: NM+0.25mM 5-FOA
- Wells 11, 24, 37, 50, 63: NM+0.5mM 5-FOA
- Wells 12, 26, 38, 51, 64: NM+1mM 5-FOA
- Wells 13, 27, 39, 52, 65: NM+2.5mM 5-FOA
- Wells 1-13: empty
- Wells 14-26: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 (6)
- Wells 27-39: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 (9)
- Wells 40-52: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 +Zif268 spec (1)
- Wells 53-65: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 +Zif268 spec (2)
Team ZF
Midiprep of Zif268 and CB Bottom Plasmids
We performed a midi-prep on the Zif268 (whole) and CB Bottom plasmids. Zif268 was eluted in 100 ul of EB, while CB Bottom was eluted in 75 ul.
- Zif had a final concentration of 261.45 ng/ul and a 260/280 of 1.90.
- CB Bottom had a final concentration of 237.93 ng/ul and a 260/280 1.91.
Results of Cross Junction PCR of 2 random colonies from ligation
After running an e-gel we saw the expected 1.4 kb band. (The gel image is not available, because we were unable to get the software running at the time). We decided to proceed with sending 24 colonies out for sequencing.
Send out 24 colonies for sequencing
We performed a cross-junction PCR on the following colonies to send them out for sequencing:
- 1B, 1C, 1D, 1E, 1F, 1G, 1H, 2A, 3C, 3D, 3E, 3F, 3G, 3H, 4A, 4B, 10A, 10B, 10C, 10D, 10E, 10F, 10G, 10H
We picked cultures that did not have sequencing results from our previous attempt. A 1 ul of a 1:10 dilution of each culture was used as template in the PCR. We used the Junction Reverse primer.
Isothermal assembly and transformation of GFP reporter
We ran our PCR product from yesterday on a 1% agarose gel. We got an appropriate band at approximately 700 bp. (Unfortunately, we could not get the imaging software to work at the time, so no image is available)
We performed a gel extraction on our GFP PCR product, with the following yields:
- 114.90 ng/ul; 260/280 of 1.89
- 92.02 ng/ul; 260/280 of 1.94
We then performed an isothermal assembly, using the following recipe:
- 0.91 ul Zif 268 (backbone)
- 0.27 ul GFP
- 1.32 ul water
- 7.5 ul isothermal assembly mix
We transformed using ChemComp cells, plating 10 and 150 ul.
CB Bottom Digestion
We digested our CB Bottom plasmid, using our usual recipe. We ran the digestion for 2 hours at 37*, 20 minutes at 65*, and then let the reaction sit at 4* overnight.August 13th
Team ZF
Sequencing Results
| Oligo | ID | Notes |
| 2 | 1B | ✓ |
| 4 | 3H | ✓ |
| 5 | 10A | ✓ |
| 5 | 1D | ✓ |
| 5 | 1E | lots of snps at beginning |
| 5 | 3G | ✓ |
| 6 | 10C | ✓ |
| 6 | 10G | ✓ |
| 6 | 3C | ✓ |
| 6 | 4A | 2 deletions at F1 |
| 7 | 10B | ✓ |
| 7 | 10D | ✓ |
| 7 | 10E | ✓ |
| 7 | 1F | ✓ |
| 10 | 3E | ✓ |
| 12 | 1C | deletion right before F1 |
| 12 | 1G | 2 deletions at end of F1 |
| 14 | 1H | insert 'T' on F1 |
| 15 | 2A | ✓ |
| 17 | 10F | ✓ |
| 17 | 10H | ✓ |
| 18 | 3D | ✓ |
| 18 | 3F | ✓ |
| 18 | 4B | ✓ |
Combined with the sequencing results from before, we are missing oligos 3, 13, 14, and 16.
Results of GFP transformation
We have colonies on both of our plates! We still need to check if they actually express GFP.
PCR purification of CB Bottom digestion product
We performed a PCR purification on the CB Bottom digestion, with a yield of 68.96 ng/ul and a 260/280 of 1.85. This will be used in a ligation in the future.August 15
TolC
There appears to be very low selection (only on the order of 1:10) for the KS:KN strain.
Wolfe
Yay!!!
Interestingly, our 5-FOA concentrations did not seem very potent: 3-AT was more effective at retarding growth. We might drop the URA3 part of our selection and instead use higher amounts of 3-AT to distinguish between true hits and false positives.
August 16
In prioritizing our work for the next two weeks, we decided to focus on three main experiments:
- Preparing the oligo pool from the chip using Sri's protocol (using qPCR to amplify the pools equally)
- Continuing the experiment of picking out a positive hit amongst negative signal
- Biobricking our expression plasmids
Eventually, the first two experiments will come together in terms of actually using the chip and figuring out how to pull out and select the positive binders.
On average we expect about three students working at a time, so we'll have these three experiments running in parallel by each student. So this means our previous teams will have to be dissolved, also since TolC will no longer be used :'(.
Each experiment in more detail:
- We need to work out the protocol with Sri in order to prepare the oligo pools for ligation into our target plasmid backbones. This will involve running a qPCR to monitor the amount of amplification so that the oligos in each pool will be amplified in equal amounts (so that we get a representative pool of oligos that is not biased to one side or the other). We will need to do this with each subpool. Futhermore, we will have to confirm that the primers that Sri provided are indeed specific enough to pull out their target subpools only and not other subpools. This is important for the tweaking of our selection system, since we will need to know whether our selection system needs to be specific enough to pick out signal from 1:10,000 or 1:100,000.
- We will need to assemble a control practice library from the 18 practice plate oligos. We will basically formulate a negative control pool with equal amounts of each of the 18 oligos, and spike this pool with varying amounts of Zif268. We will then transform our selection strain with these pools with varying amounts of Zif268, and measure how specifically we can pick a winner out amongst non-winners (so instead of a bacteria dilution, it will be a plasmid dilution). We will use 8 different concentrations:
- Positive control (all Zif268)
- 1:10
- 1:100
- 1:1,000
- 1:10,000 (this is approximately the ratio we expect for a winner amongst a pool of 9,149 non-winners for the chip subpools)
- 1:100,000
- 1:1,000,000
- Negative control (only contains the 18 practice plate oligos)
- We will start figuring out the procedure for converting our expression plasmids into Biobricks for submission. This entails locating and eliminating all of the five specified restriction sites from our biobrick part (XbaI, SpeI, NotI, PstI, EcoRI), and assembling the part into a Biobrick submission plasmid ([http://partsregistry.org/wiki/index.php/Part:pSB1C3 pSB1C3]). This may be accomplished through the use of isothermal assembly to combine the Biobrick shipping plasmid backbone with our Biobrick part (the omega subunit + zinc finger array).
Sarah
TolC
TolC selection abandoned.
Wolfe
There appears to be low selection for the Wolfe strain as well. i.e. 1:10. One possibility is that the wrong strain was picked, the other. In the absence of the 96 well plate, we will use the culture tube, and possibly the 100 well, plate reader plate.
Team ZF
GFP reporter strain
We looked at the plate of GFP reporter that grew colonies and had the correct Shine-Dalgarno RBS with the correct spacer, putting it over the blue light of an e-gel box. We have glowing colonies! Only about seven of the ~250 colonies glowed, which may be due to an extra leaky promoter. We then picked a glowing colony and put it in the warm room to grow overnight in preparation for miniprep.
Control library assembly
We picked 24 colonies from our plate which was transformed with the library mix of the 18 practice plate oligos and grew them up in LB/spec for miniprep tomorrow.August 17th
Justin
Miniprep of negative control library colonies
Today I miniprepped the 24 colonies that were picked and grown to saturation last night. These 24 colonies should contain a good number of the 18 practice plate plasmids within them. The minipreps were very successful for these strains, yielding concentrations from ~30-60 ng/ul. Nida ran a junction PCR on the 24 miniprepped products, and we sent them out for sequencing in order to determine which of the 18 practice plate oligos are contained within these 24 colonies. The yields of the minipreps can be seen below:
Selection plasmid concentrations
In order to proceed with the experiment #2 outlined yesterday, Nida and I combined the miniprepped products (everything except for sample 6, which had very low miniprep yields for some reason) in equal molar ratios according to their concentrations. This constitutes our negative control library, containing complete zinc fingers targeted towards the CB bottom target, but not towards the Zif268 binding site.
I then took the negative control library and spiked it with varying amounts of Zif268 whole plasmid, yielding mixes that contained in total 500 ng of plasmid. Thus, for the positive and negative plasmid controls, they each contained 500 ng of Zif268 and 500 ng of negative control library, respectively. For the 1:10 dilution, there were 50 ng of Zif268 and 450 ng of negative control, and so on. The Zif268 plasmid that was used originated from the midiprep that Nida performed last Friday.
The 8 mixtures outlined yesterday were then transformed into the EcNR2 selection strain, and plated on spec plates. This will simply select for bacteria that contain plasmid, but not specifically for those that contain Zif268 plasmid.
To see the volumes used for creating the plasmid dilutions, see the following file: Practice Library Assembly
Chip library assembly
PCR of Chip Oligos
With Sri and Dan’s guidance, I performed a qPCR on the chip oligo's using the protocol outlined in the supplementary section of Sri's paper. I performed six reactions using the appropriate primer in each case (CB top, CB bottom, FH top, FH bottom, Myc 981, Myc 198). I used the following recipe for the PCR (total volume 200 ul):
- 100 ul Sybr Green master mix
- 200 nm primer (4 ul each of 10 um primer)
- 2 ul chip
- 90 ul water
I ran the reactions on the thermocycler, while standing by to see the graphs being produced, using the following program:
- 95* for 1 minute
- Initially, the reactions occur at an exponential rate. During this period:
- 95* for 10 seconds
- 62* for 30 seconds
- Once the reaction began to plateau, I stopped the reaction. Prior to this point all the oligos were being amplified equally. Once the plateau occurs, amplication occurs at varying rates. This took about 12 cycles total.
FH Top samples had slightly less volume, and plateaued earlier. This may have occurred due to too little water or master mix being added.
I performed a PCR purification on the PCR product at got the following yields:
Name Conc. 260/280 CB Top: 18.2 1.83 CB Bottom: 25.1 1.78 FH Top: 20.3 20.3 FH Bottom: 21.4 21.4 Myc 981 26.1 1.73 Myc 198 19.7 1.84
In order to confirm the nanodrop readings, I ran 1 ul of each sample on a 4% e-gel. We ran a low range quanitative DNA ladder, at varying dilutions.
August 18th
Justin
Transformation results
The plates of transformed bacteria yesterday were uneven in their efficiencies because some of the plates had a low time constant for the electroporation (around 4.8) when a good time constant was around 5.8. We got excellent efficiency in the transformation for Zif268 alone, but for the plasmid mixes that included the negative control library, we got poor time constants. We still got colonies, but instead of getting a lawn of bacteria there were approximately 150 discrete colonies on the plate.
To get around this, Noah picked colonies from the plates and inoculated fresh LB/spec with them, and let them grow to saturation over the course of the day. We used these resulting saturated cultures in our selection plates later in the day.
Pre-transformation library selection plates
Today, Noah and I prepared selection plates to test our ability to distinguish a positive hit among negative controls in varying proportions. The pre-transformation aspect refers to the fact that it is the Zif268 plasmid that is diluted into the negative control plasmids before transformation, rather than diluting Zif268 bacteria into the negative control bacteria after transformation (this is analogous to how it will be done with the chip). Using the negative control library assembled from yesterday, we created a 96-well plate with the following dimensions.
Going horizontally (each column):
- 1: Positive control (only Zif268)
- 2: Positive control (replicate of column 1)
- 3: 10^-1
- 4: 10^-2
- 5: 10^-3
- 6: 10^-4
- 7: 10^-5
- 8: 10^-6
- 9: Negative plasmid control (only negative control library)
- 10: Negative plasmid control (replicate of column 9)
- 11: No plasmid control (Wolfe ECNR2 strain without any plasmid)
- 12: No plasmid control (replicate of column 12)
Going vertically (each row):
- A: LB + carb
- B: NM + his (0.1%)
- C: NM
- D: NM (replicate of row C)
- E: NM + 1 mM 3-AT
- F: NM + 3 mM 3-AT
- G: NM + 10 mM 3-AT
- H: NM + 25 mM 3-AT
We left this in the plate reader overnight.
Selection plate seeding protocol
Notes: Each well contained 150 ul of liquid, and a 1/100 dilution of bacteria at an OD of 3 (so this would be adding 1.5 ul of bacteria). The bacteria were grown up in LB to saturation and were washed in NM twice. This was accomplished by the following steps:
- Take the equivalent of 800 ul of 1.5 OD bacteria into a tube.
- Centrifuge the bacteria into a pellet (max speed for 1 min)
- Pipette off as much LB as possible, using a 1000 ul pipet first and then switching to a 200 ul pipet.
- Resuspend the bacteria pellet in 500 ul of NM (1st wash).
- Recentrifuge into pellet.
- Repeat steps 3 and 4 (2nd wash).
- Resuspend in 400 ul of NM (this gets it to an OD of 3).
- The bacteria sample is now ready to be used.
GFP reporter plasmid miniprep
Today, Nida and I miniprepped 6 ml of GFP reporter cells, consolidating the equivalent of 2 minipreps into one column. However, the yields and purities were pretty terrible, with yields of 7 and 11 ng/ul and purities that were much greater than 2. This is likely to be caused by the fact that the cultures that we miniprepped were prepared by spiking 3 ml of LB/spec with 1 ml of saturated culture in an attempt to grow up a lot of culture in a short period of time. There was likely a lot of cell lysis and general cell debris that hindered the miniprep, causing it to give such terrible yields.
We inoculated 12 more ml of culture for miniprepping tomorrow morning, since we are low on time. This should hopefully give us enough culture to work with such that the miniprep will eventually give us the quantity/concentration of DNA required for transformation and/or plasmid library testing.
Chip library assembly
Digestion of library and CB bottom backbone
We digested our CB bottom library pool and CB bottom plasmid with Bsa1 using the following recipes:
- Library (40 ul total)
- 30 ul pool library (600 ng)
- 0.75 ul enzyme
- 4 ul buffer 4
- 1 ul bsa
- 4.25 ul water
- CB bottom backbone (10 ul total)
- 4.2 ul dna (1000 ng)
- 1 ul enzyme
- 1 ul buffer
- 0.25 ul bsa
- 3.55 ul water
We ran the digestion for 2 hours at 37*. Then we heat inactivated the enzyme at 65* for 20 minutes.
Phosphatase reaction
We then took the product of the digestion, and added phosphotase to prevent the pieces from re-ligating by removing the 5' phosphate. We used NEB Antarctic phosphotase. We used the following recipes:
- CB bottom backbone
- 10 ul backbone
- 1.22 ul buffer
- 1 ul phosphatase
We ran the reaction for 4.75 hours at 37*, and then heat inactivated the enzyme at 65* for 20 minutes.
We then performed a PCR clean-up on the product, with the following yields:
- Library: 9.8 ng/ul (total 294 ng)
- CB bottom: 11 ng/ul (total 330 ng)
Planning for ligation of library and backbone
We were unable to perform the ligation today. If we want to use 200 ng per ligation reaction we need 1000 ng of DNA, which is the amount we started with. We can cut some more backbone, or phosphatase the backbone I had cut earlier (conc: 69 ng/ul). We can also use 100 ng of backbone instead.
Based on the yields of the PCR clean up, as assuming 100 ng of backbone, we'll need the following amounts of insert for our ligations:
Insert:backbone Amount of insert (ul) 2:1 1 3:1 1.5 5:1 2.5 8:1 4
This is because our insert is 137 bp and our backbone is 2788 bp. For equal molar ratios, we'll need 4.9 ng (or 0.5 ul) of insert for a 1:1 insert:backbone.
We also plan on running cut backbone (no phosphotase), and phosphotased backbone as controls.August 19th
Plate reader
- Yesterday's plate reader experiment looked great! There is definitely a difference between 10^-6 dilution of Zif268 and the negative controls!
- We will duplicate this experiment in a slightly different way to validate it. Using the same media conditions, we will dilute the amount of Zif268 spec plasmid post-transformation rather than pre-transformation. (Instead of adding different amounts of plasmid to the cells being transformed, we will add different ratios of saturated EcNR2 selection strain+Zif268 spec and saturated EcNR2 selection strain+negative control plasmids.)
- For the protocol we will use in seeding the plate, see yesterday's entry.
GFP reporter Zif268 plasmid transformation
- We also want to duplicate the plate reader experiment using our GFP reporter strain, so that we can show definitively that there is more Zif268 (represented by the GFP) in one well versus another.
- Miniprep of GFP Zif268 reporter plasmid: 151.5ng/uL, 260/280=1.89
- Transformed 1.5mL EcNR2 selection strain with 1uL plasmid (time constant of 4.8)
- Recovered 1hr in LB, and then added spec
- Grew 1hr in spec and then reinoculated 30 uL into a fresh tube of LB+spec to grow overnight (seeing as even with a poor time constant, 100 ul yielded over 100 colonies on the plate that Noah had tried earlier).
Chip library assembly
Adding phosphatase to CB bottom
Because we did not cut enough DNA yesterday, we decided to add phosphatase to a sample of CB bottom backbone we had cut before. This sample, which had already been PCR purified and eluted in 30 ul of EB, had a concentration of 69 ng/ul according to the nanodrop.
In order to make sure the sample was cut appropriately, we ran it and a sample of the CB bottom we cut yesterday on a 4% e-gel.
The band was much more intense for the old sample, and it looked like it had a concentration of between 40 and 100 ng/ul. We decided to perform the reaction on 20 ul of the cut sample (for approximately 1380 ng of DNA) and used the following recipe:
- 20 ul DNA
- 1 ul phosphatase
- 2.33 ul phosphatase reaction buffer.
We incubated the reaction at 37* for 1 hour 10 minutes, and heat deactivated the enzyme by heating it at 65* for 20 minutes.
We then PCR purified the sample, eluting in 30 ul of EB, and got a yield of 41.7 ng/ul, meaning we had a total of 1251 ng of backbone, which was more than enough for our ligation.
Ligation of CB bottom and library inserts
We performed a ligation using 100 ng of backbone and insert to backbone ratios of 2:1, 3:1, 5:1, and 8:1. We also ligated the cut plasmid and the phosphotased plasmid as controls. We used the following recipe for our ligation:
- 2.4 ul backbone (100 ng)
- insert:
- 2:1 -> 1 ul
- 3:1 -> 1.5 ul
- 5:1 -> 2.5 ul
- 8:1 -> 4 ul
- 2 ul 10x ligation buffer
- 1 ul ligase
- water to bring the total volume to 20 ul
August 20th
Plate reader for ECNR2 Wolfe
- Yesterday's plate reader results: the trends are ok but the graphs are messy, probably because of condensation on the lid.
- This experiment is to figure out the appropriate concentration of IPTG, with varying concentrations of 3-AT.
- the concentrations are
- IPTG concentrations from 0,10 and 100mM (columns 1-4, 5-8, 9-12)
- 3-AT concentrations from 1,3,10 and 25mM (rows E,F,G,H)
- LB (A), NM+histidine (B), NM (C & D)
- The four strains are
- EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 + no plasmid (columns 1, 5, 9)
- EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 + CB_bottom plasmid (negative control, library plasmid) (columns 2, 6, 10)
- EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 + zif268 spec plasmid (selection strain with the matching ZFB and ZFP)(columns 3, 7, 11)
- EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 + (zif268+GFP) spec plasmid (selection strain with the matching ZFB and ZFP, should fluoresce)(columns 4, 8, 12)
- We will take one fluorescence reading before placing it in the plate reader, and one after we finish it.
- In addition to the plate reader, we are curious to see how IPTG affects fluorescence levels. We made LB+spec plates with 0, 6.25uM, 25uM, and 100uM concentrations of IPTG
- Each plate was spread with 1uL of EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 + (zif268+GFP) and left overnight at 30C
- Tomorrow we can compare fluorescence levels between the different IPTG concentrations.
Chip library assembly
Transformation
We ran the ligation products through a min-elute column, eluting in 10 ul of water, with the following yields:
- Cut: 22.4 ng/ul
- Phosphotased: 24.2 ng/ul
- 2 to 1: 25 ng/ul
- 3 to 1: 25.9 ng/ul
- 5 to 1: 20.1 ng/ul
- 8 to 1: 20.9 ng/ul
Later we ran a transformation, using the turbo competent cells protocol. We used 100 ng of ligation product for the transformation:
- cut: 4.46 ul
- phosphotased: 4.13 ul
- 2 to 1: 4 ul
- 3 to 1: 3.86 ul
- 5 to 1: 4.98 ul
- 8 to 1: 4.78 ul
August 21st
Chip library assembly
Results of library transformation
Colonies grew on all the plates. However there were many colonies on the two control plates as well. There was a small amount of uncut (or nicked) plasmid according to the gel ran earlier, so this may be a result of that. Strangely, there were more colonies on the two control plates, than on the 2:1 and 3:1 10 ul plates. This does not make sense because one would expect that the controls demonstrate the amount of background, and the other plates should have more colonies on top of that in addition to the background noise.
The 5:1 plate had noticably more colonies than the others, including the control plates, making it the ideal insert:backbone for ligation.
Minipreps and glycerol stocks
We performed a 4 2 mL minipreps on the 5:1 culture, eluting in 30 ul of EB, and got the following yields:
- 1: 86.5 ng/ul
- 2: 94.0 ng/ul
- 3: 90.8 ng/ul
- 4: 94.2 ng/ul
We also made 4 glyercol stocks of the 5:1 culture, using 333 ul of 55% glycerol and 1500 ul culture.
Cross junction PCR of library colonies
In order to see whether our colonies actually had the insert, we decided to run a cross junction PCR on 9 colonies and the CB bottom plasmid (as a control). We diluted each colony in 10 ul of water and used that as our template. We had a 12.9 ul reaction volume.
August 22nd
Zif268-GFP results
- IPTG spec plates: the plates with IPTG did have more bacterial growth (which might just be a plating error) but none of the plates fluoresced.
- Our endpoint fluorescence reads on the plate reader did not show any fluorescence, though this may have been due to an incorrect setup.
- Growth curves (see graphs below):
- GFP Zif strain grew in NM, showing that selection is working.
- In complete media, NM, and NM+1 or 3mM 3-AT, GFP Zif grew as well as the regular Zif268 strain. However, as 3-AT concentrations increased, GFP Zif began to grow considerably less compared to Zif268. The reason for this is unclear.
- There was no real difference between 0 and 10uM IPTG, but 100uM IPTG was still detrimental for cells containing zinc finger expression plasmids.
Wolfe system biobrick
- One of our biobricks will be the Wolfe selection system (ZFB, wp, his3, ura3). We had started putting this on pSB4K5, but a PCR test of our ligation showed that none of the plasmids had the insert.
- picked 23 colonies, 10uL KAPA reactions, Spe1 ZFB_F and Xba1 URA3_R primers, 65 C annealing and 90 sec elongation
- used insert containing Spe1 and Xba1 sites as positive control
- We will not continue putting the system on pSB4K5 and instead will focus on inserting it into the pSB1C3 shipping plasmid. We ordered the proper primers and quikchange oligos (removing two Pst1 sites) to complete the system.
Chip library assembly
Cross junction PCR: Take 2
We picked 9 more colonies from the 1 ul 5:1 plate, diluted them in 50 ul water, and used 0.5 ul as our template in a cross junction PCR. We also ran the reaction on CB bottom as a control. We followed the usual cross junction protocol with the following changes:
- Initial denaturation: 6 minutes at 95* (instead of 5 minutes)
- Final extension: 10 minutes at 72* (instead of 5 minutes)
It looks like none of the colonies contain an insert. In order to try and figure out what went wrong, we ran the uncut CB bottom plasmid, cut plasmid, and cut and phosphatased plasmid on a 1% e-gel.
The gel looked extremely strange, and the ladder was not present. We tried it again, this time running uncut plasmid, new phosphatased plasmid, and old phosphatased plasmid.
The gel isn't perferct, but it looks like there was indeed uncut plasmid present after digestion in the sample used for ligation.
Digestion of CB bottom
We decided to cut more CB bottom, and try a second ligation, only using a 5 to 1 backbone to insert ratio for the ligation. We also decided to do a double digestion with both Bsa1 and Xba1 to minimize the amount of whole plasmid left over. We used the following recipe:
- 4000 ng DNA (16.8 ul)
- 8 ul Bsa1 (40 units)
- 2 ul Xba1 (40 units)
- 10 ul 10x buffer
- 1 ul bsa
- 62.2 ul water
August 23rd
Selection plates
- We have shown that selection works in liquid media, but we would like to also be able to use plates with different 3-AT concentrations, since that would make it easier to pick individual colonies that grow successfully. To check sensitivity, we plated 1uL of saturated culture of cells transformed with either all Zif268 spec, all negative control plasmids, or a mixture (10^-1 Zif, 10^-2...10^-6) on 3mM NM+3-AT plates.
- Did not see successful growth, though this may be because plates were old. An attempt with 100uL showed growth on both positive and negative control plates, likely due to the LB the cells were plated with.
- We now have fresh plates, so we will try again tonight.
- washed 100uL of positive control (100% Zif), 10^-1, 10^-2, 10^-3, 10^-4, 10^-5, 10^-6, negative control (0% Zif)with NM (500uL) two times and plated on NM+spec
- washed 100uL of EcNR2 selection strain that has not been transformed with any plasmids and plated on NM.
Changing ZF binding site
- We are trying to use MAGE, but the palindromic structure of the weak promoter gave the oligos ridiculous secondary structure, so we don't expect this to work. Instead, we are going to use lambda red to switch out the current kan-ZFB construct in the cells with a new construct containing a tetracycline antibiotic cassette and ZF binding site. By tying the ZFB change to cell survival, we hope to have greater efficiency.
Plate reader
- Repeated post-transformation dilutions of Zif with negative control library.
Chip library
Phosphatase of newest cut backbone
We used the following recipe to perform a phosphatase reaction on the backbone we cut yesterday with Bsa1 and Xba1:
- 45 ul DNA
- 1 ul phosphatase
- 5.1 ul buffer
We ran the reaction for an hour and heat inactivated the enzyme at 65* for 20 minutes. We then performed a PCR clean up on the product, with a yield of 52.6 ng/ul.
Ligation of CB bottom and library
We used the following recipe for the ligation with the insert:
- 100 ng backbone (1.90 ul)
- 2.5 ul insert (for a 5:1 ratio of backbone to insert)
- 2 ul 10x ligation buffer
- 12.6 ul water
- 1 ul ligase
To test the background we also ligated the cut and phosphotased plasmid, using the following recipe:
- 100 ng backbone (1.90 ul)
- 2 ul 10x ligase buffer
- 1 ul ligase
- 15.1 ul water
We added everything but the enzyme, and then heated the mixture at 37* for five minutes to separate any DNA fragment that were non specifically annealing. We then let it cool down back down to room temperature before adding the ligase.
We ligated the product for 10 minutes at room temperature. Then we cleaned up the product using min-elute columns, and eluting in 10 ul of water. We had the following yields:
- With insert: 27.4 ng/ul
- Control: 20.8 ng/ul
Transformation of CB bottom+library and controls
We then ran the following transformations using turbo comp cells, adding 100 ng DNA to our cells:
- Cut backbone: 2.2 ul
- Cut and phosphatased backbone: 1.9 ul
- Cut, phosphatased, and ligated backbone: 4.81 ul
- Cut, phosphatased backbone ligated with insert: 3.65 ul
- Our old cut and phosphatased backbone (high background): 2.4 ul
August 24th
Plate reader results
- The cultures did not grow as well as they have in the past. This could be due to experimental error (the plate adapter was accidentally left in the reader overnight) or perhaps the cultures we inoculated with were too old. Another possibility is that the change from LB to NM-based media causes a metabolic shock, so we will try the plate reader experiment again with cultures grown up to saturation in NM+his (with the appropriate antibiotic).
- Summary: the dilutions did not grow as well as previous plate reader experiments had shown. See graph below for example.
Chip library
Results of yesterday's transformation
The background has significantly decreased, so it looks like adding the Xba1 helped.
- There is the possiblity that the background has artificially decreased because of adding that Xba1--that is, it went down because most of the plasmids have the Xba1 cut, and not the Bsa1 cut.
However, there is not much of a difference between the control plates and the transformed plates, so it looks like our transformation did not work.
- It is possible that this occured because our Bsa1 enzyme has gone bad. We used new Bsa1 when cutting the backbone yesterday. We should try re-cutting the library with new Bsa1 and transforming with that to see it helps.
Cutting the library
Ligation and transformation
Selection plates
- The plates put after 16 hours in the incubator have a lawn of very small colonies growing very slowly. The selection with no plasmid is not growing on NM, as expected, but we do see colonies on the negative control library plate. However, there are clearly a lot more cells on the Zif268 positive control, and the dilutions seem to have colony amounts in between or similar to the negative control library plate.
- We will try several different variations:
- plate 1uL of each saturated culture on NM+spec (or plain NM in the case of the selection strain without plasmid)
- plate 100uL of each washed saturated culture on NM+10mM 3-AT
August 25th
Selection plates and cultures
- 1uL NM plates: not much is growing on even the Zif268 control. 1uL may not be enough to plate in minimal media. We will let them continue growing to see if anything shows up.
- 100uL 10mM 3-AT plates: looks very promising! Zif268 has lots of small colonies, and the amount of growth steps down through the dilutions. The negative control library shows practically no growth at all. We will also keep these in the incubator to see if that helps makes the differences even clearer.
- NM+his liquid cultures: none of them are as saturated as we would like, so we'll leave them in longer.
Chip library
Results of transoformation
- We got colonies! We did not run a second background test yesterday, but based on the previous background plate (which contained the same backbone used in this reaction) most of our colonies should have the insert.
- The best plate was the 2:1 insert to backbone ligation. The 8:1 reaction had a similar number of colonies, as well. The 3:1 and 5:1 plates had far fewer.
We performed 4 2 mL minipreps (with yields of approximately 100 ng/ul for all) on the 2:1 culture and made 4 1.5 mL glycerol stocks.
Cross junction check of library colonies
To ensure that our plates actually had the insert, we ran a cross junction PCR on 8 colonies (diluting in 50 ul water). We also performed the cross junction PCR on some whole CB bottom plasmid, to see if there is a approxmiately 80 bp difference in the gel.
We used the following recipe for the reaction:
- 12.5 ul water
- 10 ul kapa
- 0.75 ul forward primer
- 0.75 ul reverse primer
- 0.5 ul template
gel
The expected product size if there is an insert is about 1500 bp, while the product without the insert is about 1400 bp. It is difficult to see whether there is a difference in the sizes of the bands.
We also sent the samples out for sequencing (with the reverse sequencing primer), to confirm whether or not our ligation was successful. If it is, then we will send out 96 samples for sequencing.August 26th
Biobricks
Zinc finger expression plasmid digests
- Yesterday's digestions with LacI and LacIq did not work, so we tried again using a slightly different protocol from [http://www.neb.com/nebecomm/products/protocol445.asp New England Biolab's Biobrick kit instructions].
- Got the same odd doublet again
- Digested one part with EcoR1 and one part with Spe1 to see if one of the enzymes would give us the expected ~50 bp fragment, but this also didn't work
- We have concluded that there must be something wrong with the LacI and LacIq sequences. Possibly one of the restriction sites is missing. We will abandon this version of the biobrick for the time being.
- pSB1C3 shipping plasmid: 1st PCR cleanup failed, so we redigested using the [http://www.neb.com/nebecomm/products/protocol445.asp New England Biolab's Biobrick kit instructions]. Minelute produced a yield of 30 ng/uL, so a lot of DNA was lost but the concentration was still workable.
- Zinc finger + omega subunit: Mineulte produced 77.8ng/uL
ZFB-hisura biobrick digests
- PCR to add restriction sites to insert:
- EcoXba ZFB_F and PstNotSpe URA_R primers; 10ng of Spe-ZFB-hisura-Xba as template; 25uL KAPA reactions
- 60C annealing and 90 sec elongation
- Minelute of 4 reactions combined = 616 ng/uL
- Digested with EcoR1 and Pst1 following NE Biolabs protocol
- Minelute: 65ng/uL
Ligation
- Followed [http://www.neb.com/nebecomm/products/protocol446.asp New England Biolab's Biobrick ligation protocol] for 3 conditions:
- pSB1C3 and ZF+omega
- pSB1C3 and ZFB-hisura
- pSB1C3 self-ligation control
- Transformed into chemcomp cells
- Plated on LB+chloramphenicol
Selection strain and zinc finger library
- Lambda red to change the binding site worked for all the sites! All had colonies growing in tet, and sequencing confirmed that the binding site was changed.
- All were grown up in LB+tet and glycerol stocks were made
- Since the CB bottom zinc finger library has been sequence verified, we will go ahead and transform it into the corresponding selection strain
- transformed with 100ng of library plasmids
- recovered for 1hr after electroporation
- plated 10uL on LB+spec, 10uL and 100uL on NM+spec, and reinoculated 1mL of transformation culture into 20mL of LB+spec
Plate reader
- We repeated the zif268 dilution experiment using cultures that had been grown up to saturation in NM+his
- Empty selection strain, selection strain+Zif268, and selection strain+negative control library cultures were spun down, washed twice with NM, and resuspended to make and OD of 3. 1.5uL was used to inoculate each well.
- Media the same as the past week's plate reader experiments.
Chip library
Sequencing results
====Sending out a plate for sequencing====August 27th
Plate reader results
- Looks great! Not all of the wells grew perfectly (perhaps a loading error?) but there is a huge difference between 10^-6 dilution and the negative controls.
September 5th
CB Bottom Library Preliminary Sequencing Results
Recap of what has happened:
After the CB bottom library was transformed into (turbocomp or selection strain?) cells, 94 colonies were picked off of the plate after transformation. These colonies were then sequenced, with the eventual objective of determining several factors about our transformed library:
- How much spread does our library cover? Theoretically, the 94 colonies should be evenly spread throughout the 9,150 chip sequences, and the likelihood of a given sequence being represented twice is very low.
- Additionally, are the backbones being represented after transformation in the same ratios as they are on the chip?
- What are the chip error rates? The process of synthesizing the chip is error-prone, but we do not know what percentage of the oligos on the chip contain errors. These errors can be SNPs or frameshifts from insertions/deletions.
Currently, the sequencing files are all located under sequences/CB bottom insert seq 08292011.
29 sequences were analyzed by hand by importing the sequence files into SeqBuilder and manually identifying the position of the F1 motif based on its translation.
Out of these 29 sequences, 6 showed a frameshift mutation, causing the F1 and F2/F3 to be expressed in different reading frames. This indicates that based on these results, roughly 20% of the oligos on the chip contain frameshift mutations. This may be ok, due to the remaining 80% being available. The mutated oligos will simply be dropped out of the pool during selection due to creation of a nonfunctional protein.
Furthermore, 2 of the 29 sequences contained SNPs. One of them changed the F in "FQCRICMRN" to an S, while the other SNP introduced an early stop codon into the zinc finger, also making it non-functional.
Out of the 94 colonies that were sequenced, 20 of the traces were sufficiently poor quality that no F1 was identified.
For the future, we will need to create a script that will compare the sequenced results to the actual chip database. This can be accomplished with the following steps:
- Perform restriction cuts on the chip in silico, that is, manually edit out the primer tag/type II binding site, leaving only the part that encodes the F1 finger. This will be the dictionary that we will use to check all of our sequences against.
- Parse the sequencing data by identifying the part that codes for F1. This can be accomplished with regular expressions. One would need to identify the end of the omega/F1 linker (the last 30 or so nucleotides) and the beginning of the F1/F2 linker (the first 30 nucleotides). Everything in between can be assumed to be F1 material.
- The program will then check to see if the F1 that is parsed from the sequencing data exists in the dictionary. If it does, that is great. If it does not, it will do one of two things:
- Perform a modulus 3 operation to see if there was a frameshift. If the F1 is not a multiple of 3, it can be assumed that a frameshift occured.
- Look for the closest entry in the dictionary to the errant sequence (something off by 1 nucleotide, for example)
September 7
File Processing for Library Sequencing
Preliminary processing of the library sequences consists of agreggating the 94 individual fasta files into one multifasta file (which will be easier to work with) and taking the reverse complements of all the sequences (since we used a reverse primer for sequencing, the 100_bp_junction primer).
A Python script for converting fasta into multifasta can be found here: [http://sourceforge.net/projects/f2m2f/], and a website for converting a multifasta file into its reverse complement can be found here: [http://www.ualberta.ca/~stothard/javascript/rev_comp.html].
The reverse complement multifasta file for the 94 sequences thus far can be found in the dropbox under sequences/CB bottom insert seq 08292011 with the name CB bottom seq multifasta RC.fasta.
Remaining efforts include parsing the file to extract the F1 subunits, which will require either removing the newline characters and using regex or by using a regex that can recognize sequences across multiple lines (i.e. regex that recognizes sequences with newline characters randomly in there). The chip dictionary still needs to be built and the code for comparing our sequences with the dictionary also still needs to be written.September 8th
- Biobricks:
- The plates looked good: the plates with the omega-ZF and ZFB-his3-ura3 inserts had many more colonies than the pSB1C3 self-ligation control.
- Chose 4 colonies from each biobrick and will grow them overnight. Tomorrow they will be miniprepped and sent out for sequencing using the standard primers on the backbone. (This should cover the entire insert for omega-ZF and most of the insert for ZFB-his3-ura3.)
- Selection strains:
- Streaked out fresh plates from the glycerol stocks of the EcNR2 selection strain with the various zinc finger binding sites. We will hopefully use these when the other libraries are assembled (and we can use the CB bottom colonies for a fresh transformation with its library).
September 11th
- CB bottom ZFB strain:
- Repeated the transformation with CB bottom ZF library (100ng)
- recovered for 1 hr in LB
- plated on LB+spec to check that the transformation worked
- reinoculated 1mL of the transformation into 10mL LB+spec and let it grow for another hour
- plated 10uL (brought up to 100uL in NM) on LB+spec, NM+his+spec, NM+spec, NM+spec+1mM 3-AT, NM+spec+3mM 3-AT, NM+spec+10mM 3-AT plates
- let rest of LB+spec culture grow overnight to use tomorrow for glycerol stocks and more plating
September 12th
Library Construction
- Overnight cultures were miniprepped (yields around 30-40ng/uL) and digested following the August 22nd protocol (but with 1ug of template plasmid)
- The digested plasmid was then immediately phosphatized by adding 1uL of Antarctic phosphatase and 10uL of the phosphatase buffer and incubating for 30min at 37C followed by 5 min at 65C.
- Minelute purified the product: recovered about half of the DNA
CB bottom library
- Made glycerol stock of CB bottom strain + CB bottom library
- LB+spec plates look great, so the transformation worked! Could be small colonies on the NM-based plates, but they are very small so we will let them grow up for longer
- Also want to plate washed cells from the transformation: washed twice in NM, and then plated either the equivalent of 100uL or 10uL of cells onto LB+spec, NM+his+spec, NM+spec, NM+1mM 3AT+spec, NM+3mM 3AT+spec, NM+10mM 3AT+spec plates.
September 17th
- ZFB-his3-ura3 biobrick:
- Digested insert and pSB1C3 backbone with EcoR1 and Spe1 following NEBiolabs protocol
- Minelute: 50ng/uL and 17ng/uL respectively
- ligated according to NEBiolabs protocol (both with and without ZFB-hisura insert) and transformed into chem comp cells. Recovered for 1 hr and plated on LB+chloramphenicol
- CB bottom library:
- Retransformed CB bottom ZFB strain with CB bottom library and Zif268 as a negative control (about 100ng each)
- Plated 10uL (unwashed) of both cultures on the full range of plates. The rest of the culture was inoculated with spec and left to grow overnight.
- Library assembly:
- Ran the digested and phosphotased samples on a gel with the uncut plasmids to verify that they were cut correctly. The cut samples ran differently from the uncut, showing that the plasmids were successfully linearized.
September 19th
- PCR using VF2 and VR primers to check that biobrick assembly worked
- Results: bands were too small--insertion not successful
- repeated digestion (NEBiolabs protocol) for ZFB-hisura and pSB1C3 with EcoR1 and Spe1
September 21st
- ligated and transformed ZFB-his3-ura3 biobrick into chem comp cells using same procedure as before
- CB bottom library plates: some of the plates looked like they may have colonies, so we picked a few to grow up in LB+spec for further confirmation.
September 22nd
- Ligation plates still had very few colonies, but when I repeated the PCR with the VR and VF2 primers, it looks like one of them (#18) does have the proper insert! Culture will grow overnight and be miniprepped tomorrow for Quikchange.
- Miniprepped TolC selection strain KS to get a fresh batch of Zif268 on the spec plasmid
- 26 of the 30 CB bottom cultures grew overnight in LB+spec, so I did a PCR of the pZE23G plasmid junction (same procedure as July 12th). Zif268 acted as a positive control. It appears that most of the colonies that grew do indeed have the zinc finger expression plasmid in them--they might be novel hits!!!!
September 23rd
CB bottom hits:
Plate reader:
- Columns 1, 7: LB+spec
- Colummns 2, 8: NM+his
- Columns 3, 9: NM
- Columns 4, 10: 1mM 3-AT
- Columns 5, 11: 3mM 3-AT
- Columns 6, 12: 10mM 3-AT
- Row A: negative control (CB bottom +Zif268)(1-6) and D1 (7-12)
- Row B: D2 and C3
- Row C: B1 and B2
- Row D: B3 and B4
- Row E: B5, B6
- Row F: B7, B8
- Row G: B9, B10
- (D1-2 are from 10mM 3-AT plate, C3 from 1mM 3-AT, all Bs from 3mM 3-AT plate)
Sequencing:
- Repeat yesterday's PCR to be sent out for sequencing
Biobricks:
- Miniprepped chem comp cells with ZFB-hisura biobrick for Quikchange tomorrow.
 "
"