Team:Harvard/Project/Design
From 2011.igem.org
Overview | Design | Synthesize | Test | Zinc Finger Background | Protocols
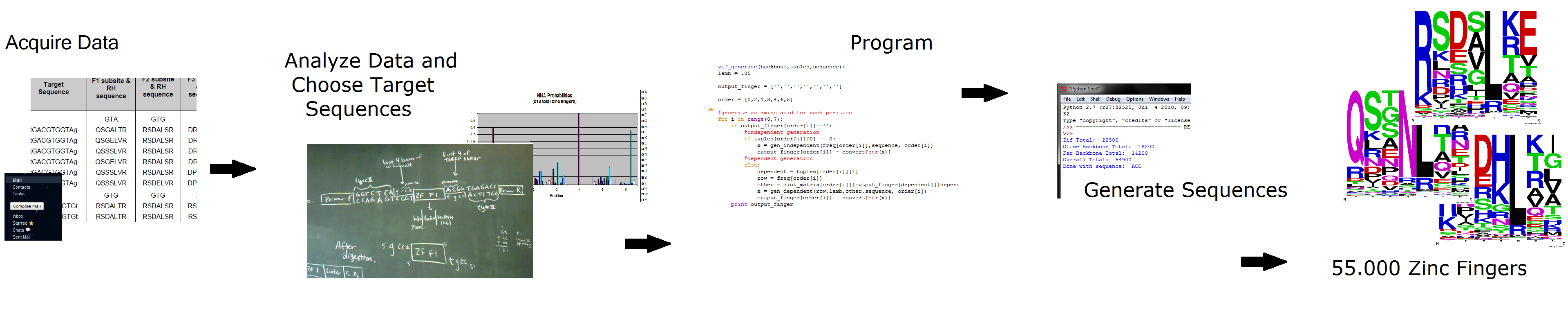
We emailed the OPEN consortium and Anton Persikov to acquire their respective databases of zinc fingers: Persikov had compiled the results of the past 10 years of zinc finger research. We then decided how to analyze this data and choose targets for new zinc fingers (details below). Then we programmed our ideas into a Python program, and generated 55,000 potential zinc fingers.
Make your own zinc finger sequences using our [http://sourceforge.net/projects/harvardigem/ generator]. Written in Python 2.7, this program is what team members created to generate 55,000 zinc finger sequences.
Contents[hide] |
Terminology
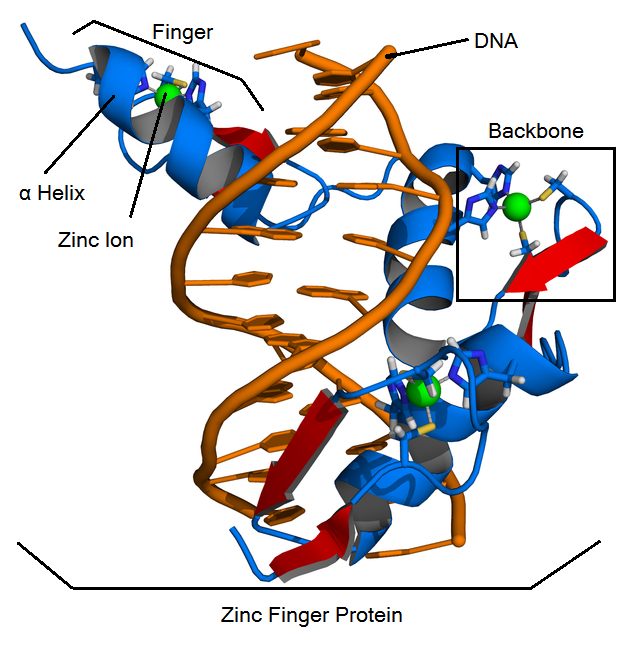
- Backbone: contains most of the amino acids of a zinc finger protein: zif268 is the most famous backbone.
- Fingers: contain a backbone and a helix, bind to a 3-base DNA triplet
- Helix: the alpha helix in a finger. It is responsible for binding to a DNA triplet. Helices are made up of 7 amino acids, and fit into a specified position in a backbone. Amino acid positions are specified by -1,1,2,3,4,5, and 6.
- Zinc finger proteins (ZFPs): arrays of three fingers that bind to 9 bases (3 triplets) of DNA.
Past Zinc Finger Designers
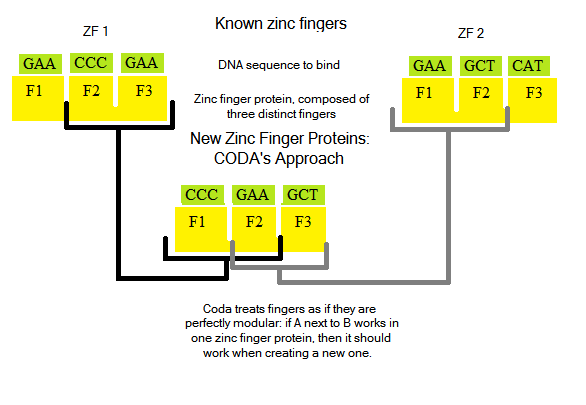
Designing new zinc finger proteins (ZFPs, which are arrays of three fingers) is not an easy task: how they bind and interact with DNA bases is not fully understood, and is an active area of research ([http://www.princeton.edu/~persikov/publications.html Persikov]). Notable past attempts to create novel ZFPs ([http://www.nature.com/nmeth/journal/v8/n1/full/nmeth0111-53.html CODA], [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2535758/ OPEN]) tried a two distinct methods: CODA took a modular approach.
OPEN took two known fingers from an array, and randomized protein sequences to try to generate a third finger to bind a new triplet:
Both techniques were successful in finding ZFPs to bind to novel DNA sequences.
Our Approach
Improving on the concept of OPEN, we decided to design ZFPs where the first two DNA triplets can be bound, but the third cannot. For example, if the sequence GTG GGA CCA can be bound but GTG GGA TGG cannot, we would use the first two fingers and generate the third. OPEN simply randomized amino acid sequences to try to create a third finger: we wrote software that uses data from known fingers to "intelligently" generate new fingers.
Selection of Target Sequences
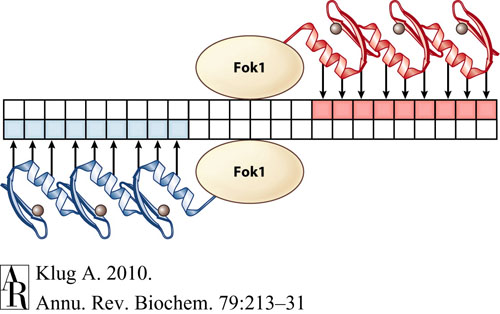
We decided to select six target DNA sequences that would serve as targets for our approach to designing custom zinc fingers. These six sequences were selected due to their potential as gene therapy targets that can be targeted with zinc finger nucleases (ZFNs), which would be capable of inserting or deleting genes with great precision. ZFNs are composed of zinc finger arrays fused to FokI type II nucleases, and they induce double strand breaks in DNA by binding to specific DNA sequences flanking the desired cut site. These sequences are on opposite DNA strands, and are separated by a spacer that is typically 5-7 bp long.
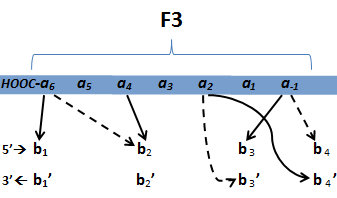
In the following image, the blue zinc finger array represents the "bottom" ZFP while the red represents the "top" ZFP, each binding to their respective 9-bp DNA sequence. Note the arrangement of the two ZFPs such that the FokI nucleases align for a double stranded DNA break.
We selected six target sequences across three genetic diseases, which include red-green colorblindness, familial hypercholesterolemia, and cancer caused by activation of the Myc oncogene. These diseases were chosen due to their monogenetic nature in which insertion or deletion of a single gene carries the potential for treatment. In terms of how we decided specifically which target sites to pursue, we followed the following steps:
- Using the CoDA tables published in the supplementary data of [http://www.nature.com/nmeth/journal/v8/n1/full/nmeth.1542.html Sander et al, 2010 in Nature Methods], we found 9-bp DNA sequences with a known F2/F3 relationship but with an unknown F1/F2 relationship. For instance, the helix sequences for GNNGNN are known (corresponding to the F2 and F3 helices, respectively), but the helix sequences for GNNCNN are unknown (corresponding to the F2 and F1 helices). In total, this 9-bp DNA sequence would be 5'-GNNGNNCNN-3'.
- In total, we defined six broad target sequences of varying "risk", based on what currently exists in the CoDA tables. For instance, an F1 triplet of GNN or TNN is much more characterized, and hence less "risky" than a triplet of ANN or CNN.
- Using the UCSD Genome Browser, we searched nucleotide sequences from 3kb - 10kb in size for the six broad target sites as defined above. For diseases that would be treated with gene insertion (i.e. colorblindness and familial hypercholesterolemia), this nucleotide sequence was located in an "empty" stretch of DNA that contained no known genes. For diseases that involved gene knockout (i.e. Myc-related cancers), the gene itself was used as a search space for ZFN binding.
- The resulting target sequences can be found in the table below:
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,403,001-153,407,000 | 3627 | GTG GGA TGG | GAA GGG ACC | RNTALQH.QSAHLKR.####### | QDGNLGR.RREHLVR.####### |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 14001 | GGC TGA GAC | GGA GTC CTG | ESGHLKR.QREHLTT.####### | QTTHLSR.DHSSLKR.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 198 | GGT GCA GGG | GGC TGA CTC | VDHHLRR.QSTTLKR.RRAHLQN | ESGHLKR.QREHLTT.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 981 | GGA GAG GGT | GGC TGG AAA | QANHLSR.RQDNLGR.TRQKLET | EKSHLTR.RREHLTI.####### |
- Green cells are our target sequences.
- The bolded DNA triplets are the targets of our variable F1 regions in our plasmid library.
- The amino acid sequences represent the corresponding binding helix sequences for each finger in the 3-finger array for each ZFP, with a "#" sign representing the unknown specific sequence that we are looking for.
Zinc Finger Binding Site Finder
To use the application that we designed to search any DNA sequence for two ZFN flanking sites, please visit our Zinc Finger Binding Site Finder. This is how we located the eight pairs of flanking sequences in the above table.
Target Site Background Information
Colorblindness (Green Opsin)
Goal: Produce functional green opsin photoreceptor proteins in the eye
Method: Insertion of functional green opsin gene (OPN1LW [http://genome.ucsc.edu/cgi-bin/hgGene?hgsid=214239613&db=hg19&hgg_gene=uc004fkb.2&hgg_chrom=chrX&hgg_start=153448084&hgg_end=153462351 1]) upstream of normal locus in patient lacking the gene
- Journal Articles
- http://www.nature.com/nature/journal/v461/n7265/abs/nature08401.html
- http://www.nejm.org/doi/full/10.1056/NEJMc0903652
- In the News
- http://www.nature.com/news/2009/090916/full/news.2009.921.html
- http://www.scientificamerican.com/podcast/episode.cfm?id=gene-therapy-cures-colorblind-monke-09-09-16
- http://www.msnbc.msn.com/id/32879284/ns/health-health_care/t/gene-therapy-fixes-color-blindness-monkeys/
- http://www.wired.com/wiredscience/2009/09/colortherapy/
Inherited High Cholesterol (Familial Hypercholesterolemia)
Goal: Produce functional LDLR protein to remove LDL cholesterol from the blood
Method: Insertion of functional LDLR gene upstream of nonfunctional allele
- http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0001429/
- http://www.genome.gov/25520184
- http://emedicine.medscape.com/article/121298-overview#a0199
Cancer (Myc Oncogene)
Goal: Knock out the oncogenic protein product and stop cancerous proliferation
Method: Targeted disruption (deletion) in mutated oncogene
- http://www.ncbi.nlm.nih.gov/gene/4609
- http://omim.org/entry/190080
Data and Analysis
OPEN provided us with a spreadsheet of ZFPs produced by their research. Anton Persikov, during his own ZFP research, has compiled a database of ZFPs from studies from 1980-2005 which he shared with us.
From these two datasets, we distilled over 3000 unique ZFPs which contained approximately 1500 unique fingers.
We analyzed this dataset for frequency (how often a given amino acid appears in a given position in the helix) and pairing (if amino acid A is in position 1, how often is amino acid B next to it).
Helix Dependencies
Amino acids do not exist in a vacuum: they must somehow be affected by amino acids around them. Besides pairing data, we realized that other interactions could be taking place, and that we needed a way to see these other relationships.
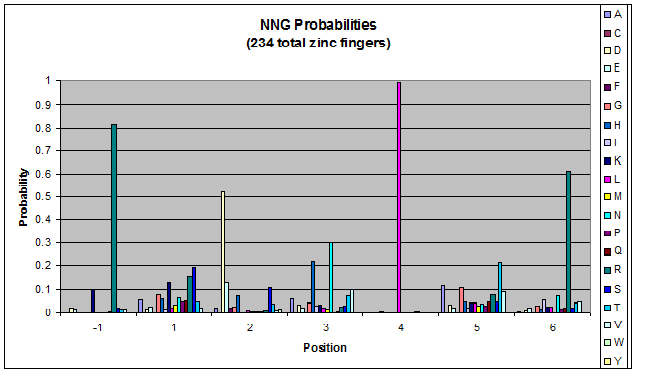
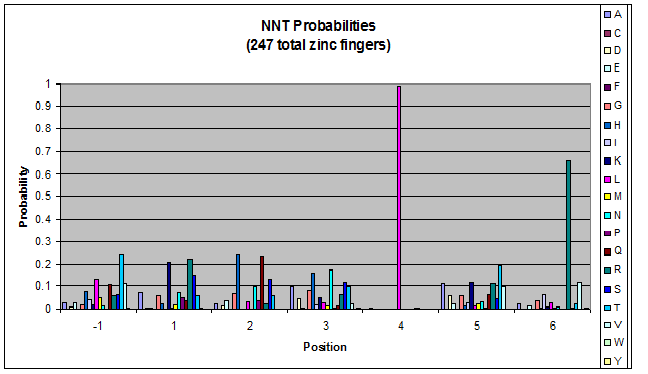
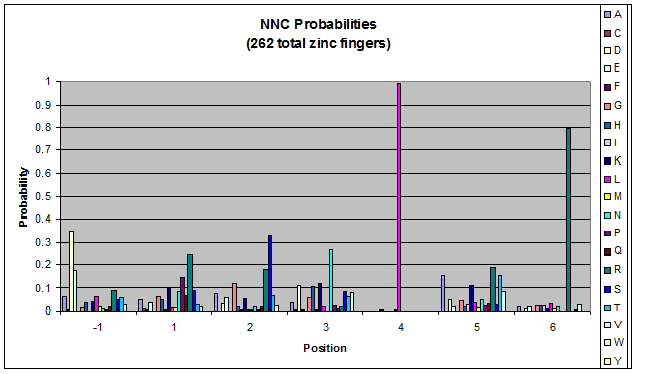
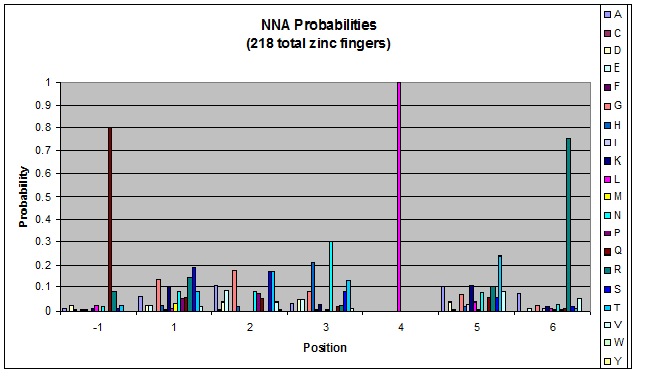
We know that the DNA bases affect the amino acid sequence, so we started looking for evidence that, for example, changing the third base (going from NNA to NNC, etc) affects position -1.
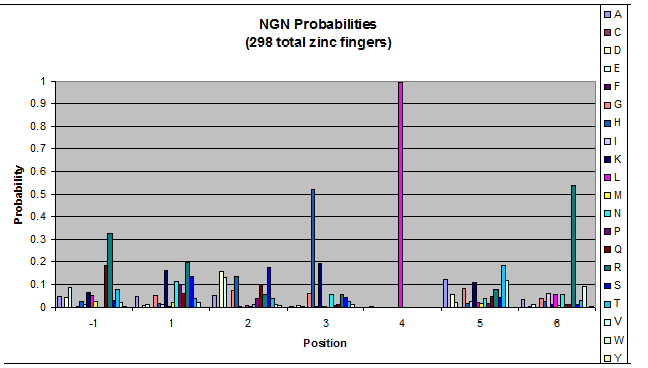
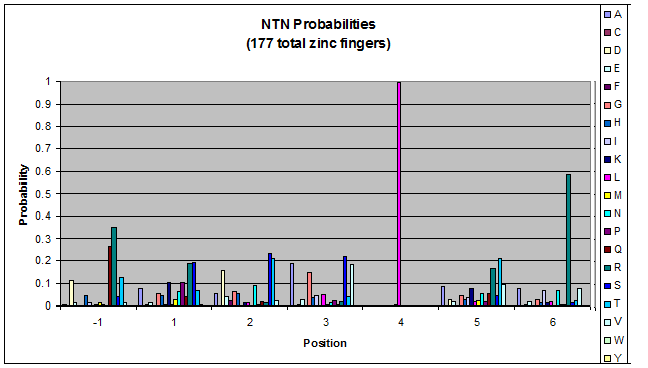
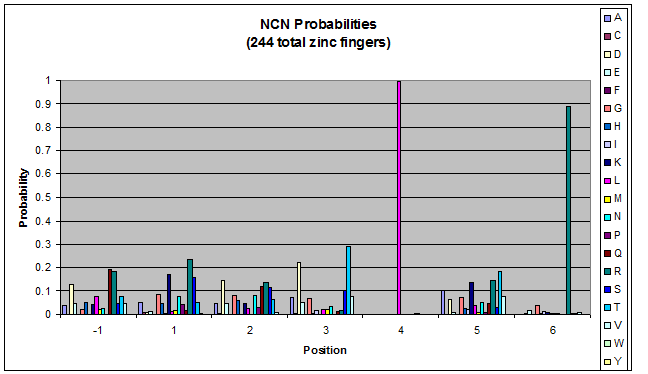
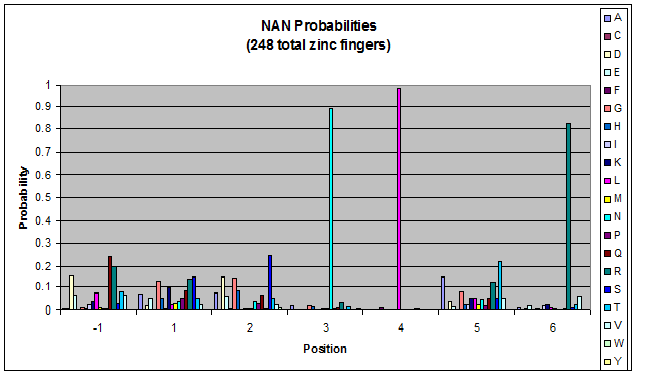
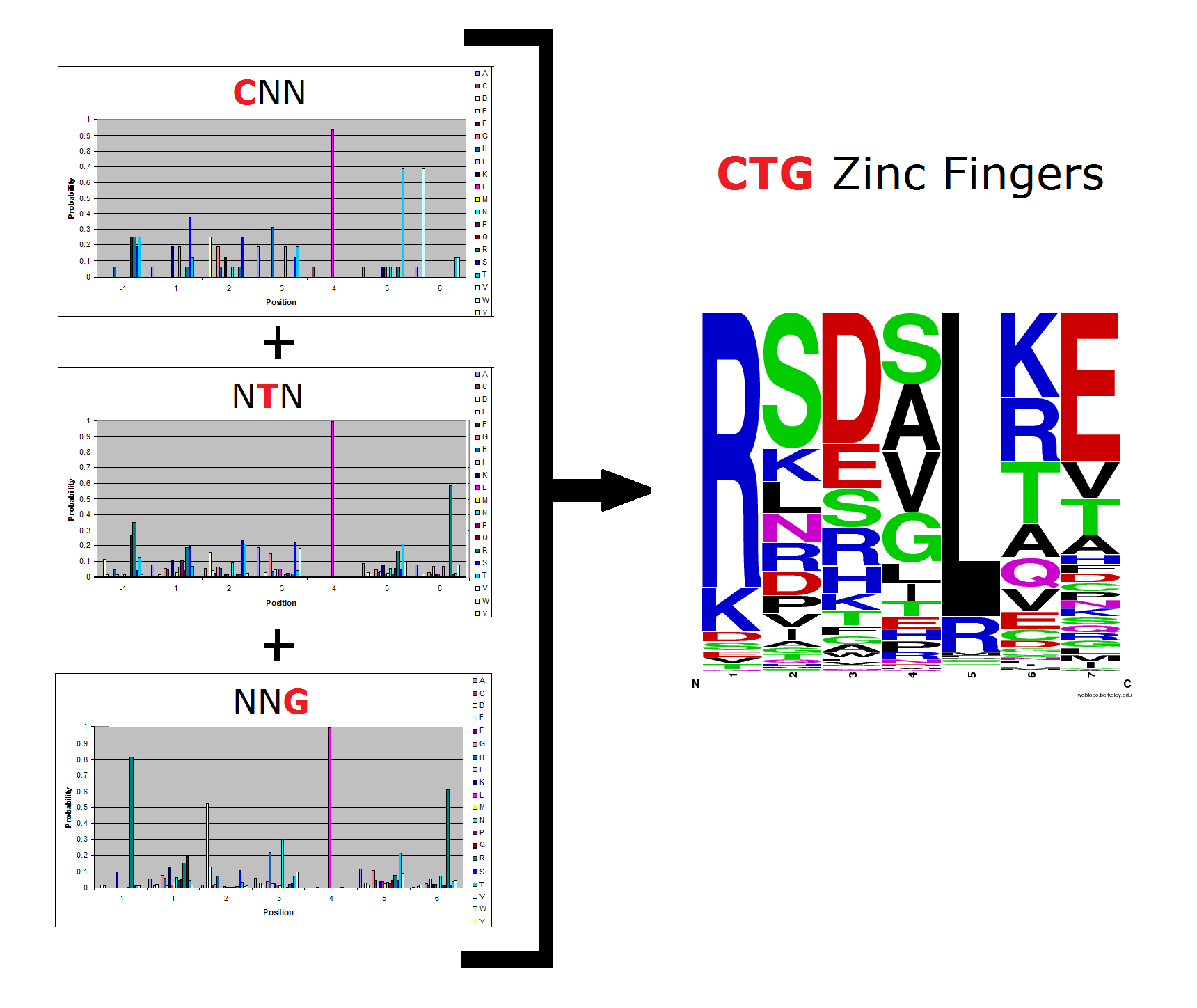
To do this, we created these graphs of the frequency of amino acids in each position, and then [http://en.wikipedia.org/wiki/Blink_comparator blinked] the graphs against each other to see what changes. We looked at the probability graphs to determine which amino acid positions on the finger's helix interact with which bases. For example, if you compare NAN to NCN, you will see a large change in the asparagine content in position 3.
We saw some interactions that are fairly well estabilished [Persikov], while others have been more recently proposed [Persikov].
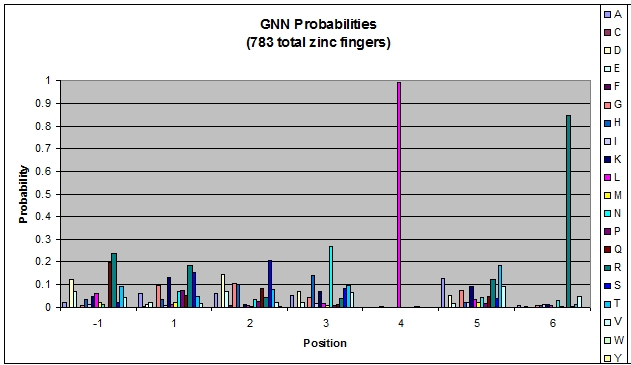
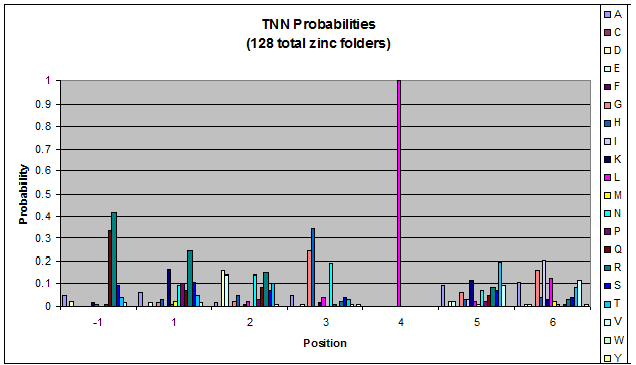
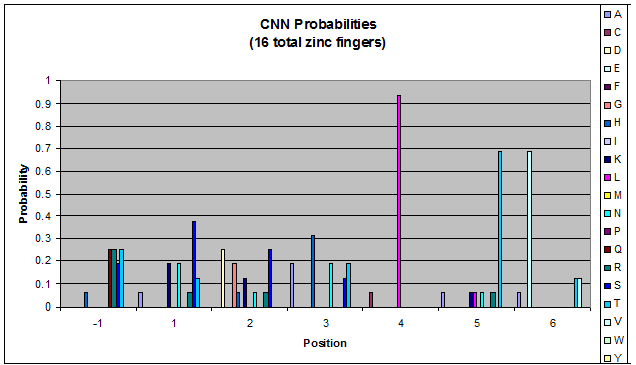
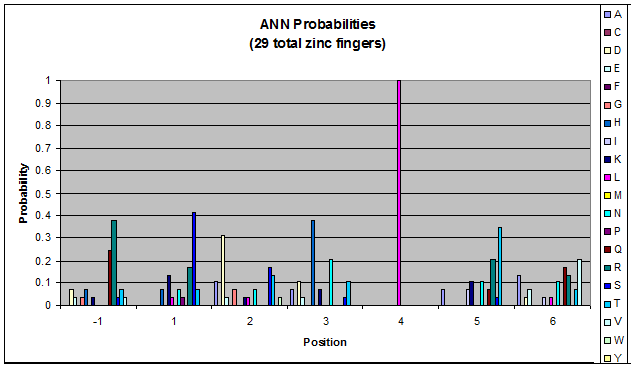
Click on the triplets on the left to compare the frequencies for various DNA triplets:
| GNN |
| TNN |
| CNN |
| ANN |
| NGN |
| NTN |
| NCN |
| NAN |
| NNG |
| NNT |
| NNC |
| NNA |
(A more rigorous way to calculate this is to calculate the entropy change as you change the amino acids in each position. But that is computationally intensive)
By doing this, we were able to see several patterns.
- xNN(Vary base 1): Amino acid 6 changes
- NxN(Vary base 2): Amino acid 3 changes
- NNx(Vary base 3): Amino acid -1 and 2(?) changes
Our program looks at dependencies between amino acids when generating sequences.
We decided on these amino acid dependencies, using both established data and patterns we saw in the OPEN data:
- -1 and 2
- 2 and 1
- 6 and 5
Because there is not much data for 'CNN' and 'ANN' sequences (with 16 and 29 known fingers that bind to each triplet, respectively), we should use pseudocounts for these sequences, so that our frequency generator is not too biased toward probabilities that may not be significant.
Programming
Overall Method: Probabilities and Randomization
Our generation program uses these amino acid frequencies as probabilities that position X contains amino acid X, given what triplet we are trying to bind. Using the dependencies we found, we change which frequency tables are used to generate the new helix. Frequency tables are built using the data from the above graphs.
See the image at right for explanation on how we turn probabilities into amino acids.
To generate one helix (7 amino acids), the program goes through the following steps:
| Step | Example for TGG |
| Generate an amino acid for position -1 (P0), using probabilities only from NNx | R _ _ _ _ _ _ |
| Taking into account the amino acid chosen for P0, generate P2, also using probabilities only from NNx | R _ S _ _ _ _ |
| Taking into account the amino acid chosen for P2, generate P1, using overall probabilities for P1 | R L S _ _ _ _ |
| Generate P3, using probabilities only from NxN | R L S H _ _ _ |
| Generate P4, using overall probabilities for P4 | R L S H L _ _ |
| Generate P6, using probabilities only from xNN | R L S H L _ M |
| Taking into account the amino acid chosen for P6, generate P5, using overall probabilities for P5 | R L S H L Q M |
These steps are based on the relationships we found from reading papers [Persikov] and studying the above frequency graphs, which were created from successful ZFPs.
The generated helix is then placed into a backbone: for example, this helix was placed in the zif268 backbone, giving a finger with a final amino acid sequence of FQCRICMRNFSRLSHLQMHIRTH.
This finger is then reverse-translated into DNA (along with the sequences for the fixed first two fingers of the ZFP) for inclusion in the chip.
Refinement: Pseudocounts
Pseudocounts are necessary for data that has small sample size - we could be missing out on working helices because a letter's frequency is 0 when it shouldn't be. For CNN and ANN, our dataset is tiny compared to GNN and TNN: CNN and ANN have around 20 datapoints while GNN has over 700. Because of this discrepancy in sample size, we must add psuedocounts to CNN and ANN in order to allow for more variation than is shown in our data.
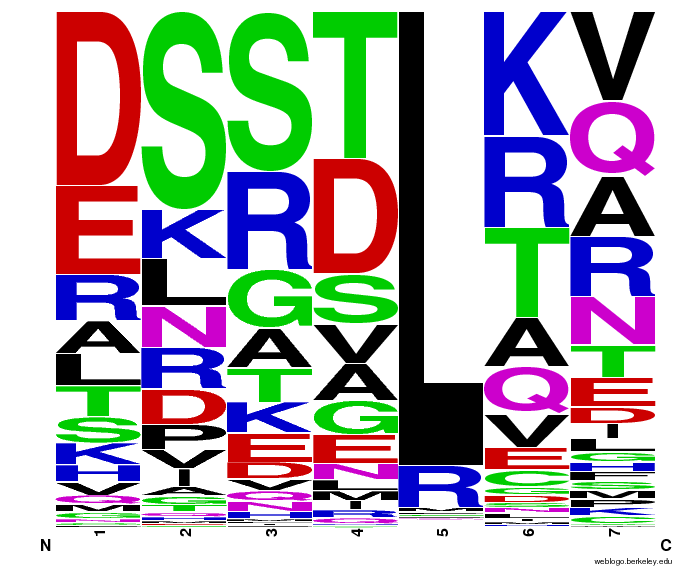
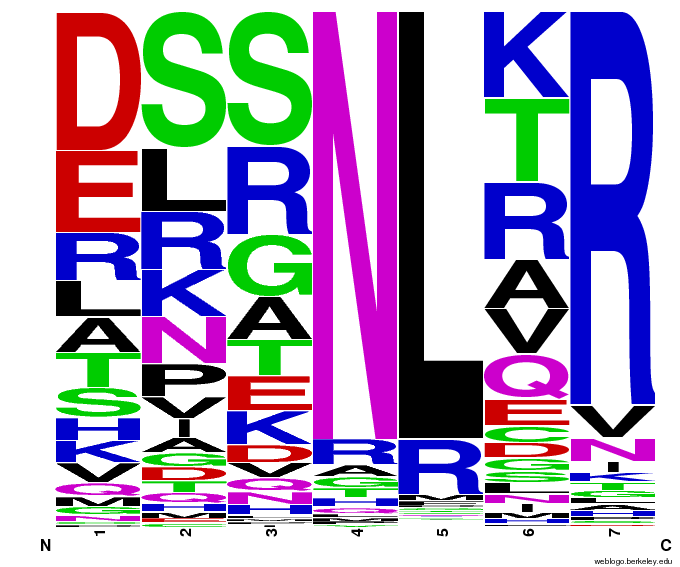
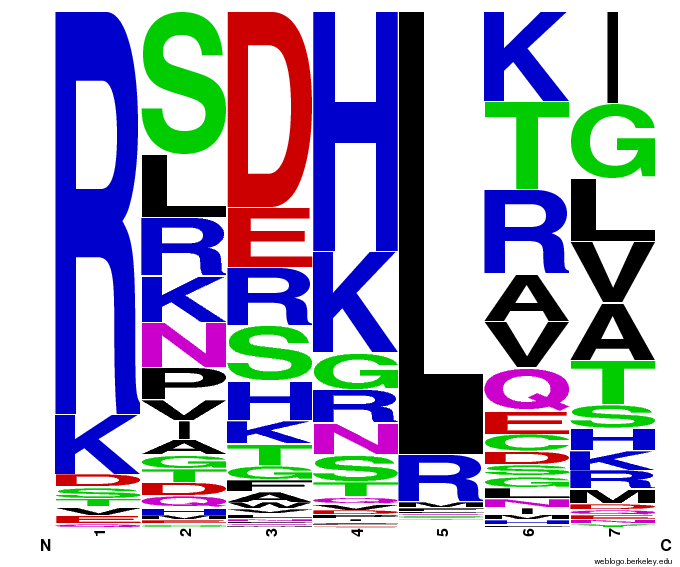
When generating helixes for CTC (because of position 6's reliance on the CNN frequencies) to test psuedocounts, we see in the created sequences the difference pseudocounts make. A psuedocount of .015 changes the frequency of any amino acid from whose frequency is 0 by bumping it up to the value of the psuedocount: ex. A = 0 becomes A = .015, giving A a 1.5% chance of being selected instead of none at all.
Visualizing our data, we get various pseudocount (psu = ) values for position 7 (which, in reality, is position 6 in the helix). The size of the letter directly corresponds to the percentage of sequences that have that letter in that position. A letter that takes up 1/3 of a column is present in that position in 33% of the helices.
Notice how psu = 0 gives only the four letters found in our CNN dataset, while psu > 0 adds in other letters, each with a small probability.
The question is how much psu to add: less means we weight our (possibly flawed) data of proven zinc fingers more. Higher psu adds more randomness (variation) to our sequences, but some (perhaps large) fraction of those sequences will not work, and take away space from the proven amino acids.
We ultimately chose psu = .015 for our software.
Results: 55,000 Possible Zinc Fingers
We made 55,000 sequences, distributed evenly among 6 DNA target triplets. That's 9150 per target.
Because our program's output changes dramatically based on the input triplet, no two sets of sequences are the same:
</body>
 "
"