Results
Contents |
NB: unless differently specified, all tests were performed in E. coli MGZ1 in M9 supplemented medium at 37°C. For the cloning of the parts, E. coli TOP10 was used.
Parts assembly
Characterization of basic modules
Characterization of promoters pTet and pLux
Characterization of enzymes AiiA and LuxI
Characterization of the efficiency of RBSs from the community collection
Identification of bacterial growth parameters
The bacterial growth curve has been modelled as a logistic curve and is represented by the following equation:
dN/dt=N*μ*(Nmax-N)/Nmax
N(0)=n0
where μ represents the growth rate of the cells (E. coli MGZ1 in M9 supplemented medium) and Nmax represents the maximum number of cells in the well. For a detailed description of the parameters, see modelling section. For details on parameters identification, see identification section.
The growth curves in all the performed experiments are measured in O.D.600. Since the N species in the model is expressed in cell number, a conversion factor has been estimated. The conversion factor KO.D.toC.F.U. has been estimated as follows.
- Two cultures C1 and C2 (MGZ1 cells) were grown in 1ml M9 medium till saturation (ON liquid culture, 37°C, 220 rpm).
- Next morning, both C1 and C2 were diluted in M9 medium with a final volume of 1ml with the following dilution factors:
- 1:1
- 1:10
- 1:100
- 1:1000
- After 1 hour, O.D.600 was measured using TECAN microplate reader (don't forget to measure a M9 sample for blanking!)
NB: from now on, cultures must be placed in ice to stop cell growth. - At the same time, proper dilution of the cultures were plated on LB agar plates.
NB: All the dilutions are performed moving 100 μl of culture in previously ice-chilled 900 μl fresch M9. 100 μl of the final dilution are plated (It still represents a 1:10 dilution!) - Plates were grown overnight and next morning C.F.U. were counted.
- C.F.U. values were corrected by the dilution factor and a linear regression (N vs O.D.600) was performed in order to evaluate the conversion factor KO.D.toC.F.U..
- KO.D.toC.F.U. was used as conversion factor to multiply the O.D.600 value of saturation in the growth curves (~0,5).
The results are summarized in the table and in the figure below:
| Culture | O.D.600 TECAN | O.D.600 Spectrophotometer | C.F.U. | Dilution Factor (10^) | N |
| C1 | 0,367950002 | 0,758004416 | 990 | 5 | 990000000 |
| C1 | 0,044649998 | 0,091982322 | 141 | 6 | 1410000000 |
| C1 | 0,004799999 | 0,009888356 | 136 | 5 | 136000000 |
| C1 | 0,000549998 | 0,001133037 | 20 | 6 | 200000000 |
| C2 | 0,54840003 | 1,129744917 | 165 | 4 | 16500000 |
| C2 | 0,058200002 | 0,119896339 | 23 | 5 | 23000000 |
| C2 | 0,008700002 | 0,017922652 | 251 | 3 | 2510000 |
| C2 | 0,000100002 | 0,000206011 | 24 | 4 | 2400000 |

The estimation of μ parameter was performed by determining the slope of the logarithmic curve of O.D.600 in exponential phase. Exponential phase was determined by visual inspection as the linear phase of the logarithmic curve of O.D.600.
The estimated parameters are summarized in the table below:
| Nmax [cell number] | μ [min-1] |
| 1*109 | 0.004925 |
The reported value of μ corresponds to a doubling time of 142 minutes.
Estimation of the spontaneous degradation of HSL in M9 medium and in cultures at different pH values
In order to estimate the spontaneous degradation rate of HSL in M9 medium and in a culture of MGZ1 cells as a function of pH, two simple tests have been performed.
Two different M9 media were prepared, one with the nominal pH (7.0) and one with pH=6.0.
These media, now named respectively M9pH 7 and M9pH 6, were added with a known concentration of HSL (100 nM) and then incubated at 37°C, 220 rpm (NB: the media were not infected with any culture but the standard growth conditions were reproduced). The amount of HSL present in the medium was assayed through the BBa_T9002 biosensor at 4 time points:
- t=0 h;
- t=1 h;
- t=2 h;
- t=4 h;
- t=8 h;
The obtained time series of HSL amounts were processed to evaluate the time constant governing the dynamic of HSL degradation, supposing an exponential decay. The results are reported in the table below:
| t1/2* [h] | γHSL** [h-1] | |
| M9pH 6 | 32 | 0.022 |
| M9pH 7 | 8 | 0.087 |
The described experiment was repeated with a MGZ1 culture in order to evaluate the effect of culture on HSL stability. The estimated values are reported in the table below:
| t1/2* [h] | γHSL** [h-1] | |
| CulturepH 6 | ∞ | 0 |
| CulturepH 7 | 19 | 0.037 |
It is evident from the reported data that the spontaneous degradation of HSL is negligible when CTRL+E is implemented in MGZ1 cells grown in M9 medium with pH=6.0 and pH=7.0.
Thus in the simulations we set γHSL=0;
Characterization of BBa_T9002 biosensor
As described in the modelling section, BioBrick BBa_T9002 is an HSL biosensor, which provides a non linear relationship between HSL input and Scell output. More precisely, the characteristic sigmoidal curve requires synthetic parameters for its accurate identification. These are the minimum and maximum values, the swtich point (i.e., the curve inflection point), and the upper and lower boundaries of linearity. This biosensor revealed greatly reliable, providing measurement repeatability and minimal experimental noise. Referring to its activation formula, the calibration curve is shown below.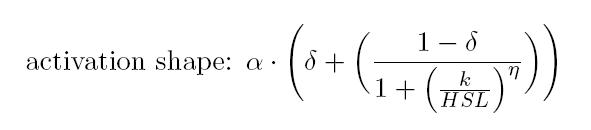
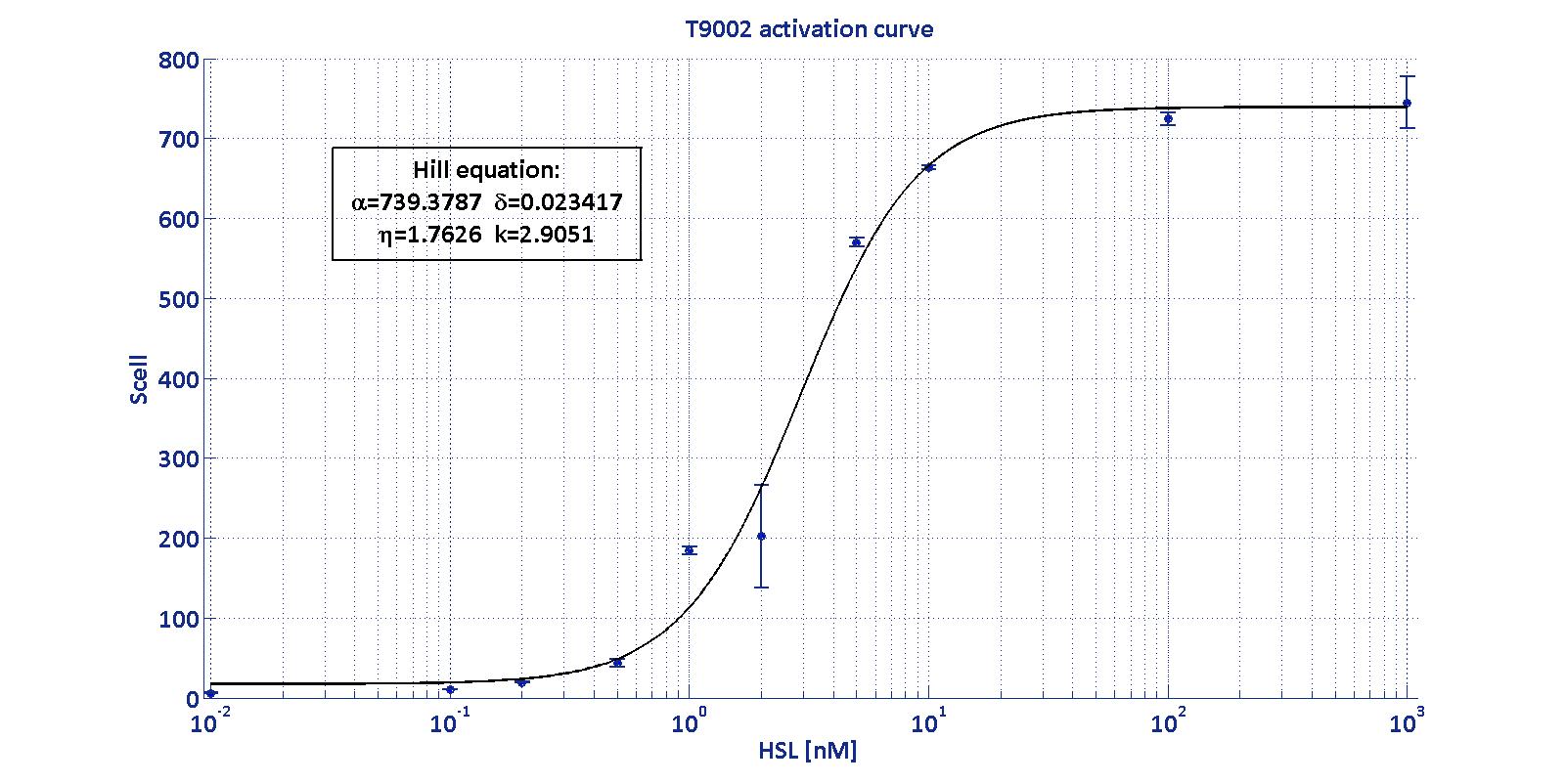
| Minimum [Scell] | Maximum [Scell] | Switch point [nM] | Lower boundary of linearity [nM] | Upper boundary of linearity [nM] |
| 17.31 | 739.4 | 1.39 | 0.38 | 5.07 |
In order to determine the threshold sensitivity of T9002 biosensor, experiments were performed with several HSL inductions minimally interspaced in the region of low detectability. These allowed to reveal that the minimum detectable HSL concentration is ??? nM.
 "
"